Summary
November 2019
Phyto-threats workshop November 13th 2019, APHA, Sand Hutton, York
Phyto-threats project team meeting
November 14th 2019, APHA, Sand Hutton, York
November 2018
Phyto-threats project team meeting
November 20th 2018, Forest Research Northern Research Station, Roslin
June 2018
June 19-20th 2018, Stoneleigh Park, Warwickshire
April 2018
Phyto-threats project team meeting
April 26th 2018, CEH, Wallingford, Oxfordshire
October 2017
Phyto-threats project team meeting
October 3rd 2017, APHA, Sand Hutton, York
Reducing Phytophthora in trade and designing effective accreditation
Phyto-threats workshop
October 4th 2017, APHA, Sand Hutton, York
June 2017
June 20-21st 2017, Stoneleigh Park, Warwickshire
May 2017
Phyto-threats project team meeting
May 4th 2017, James Hutton Institute (JHI), Invergowrie, Dundee
March 2017
Phyto-threats attendance at the 8th Meeting of the International Union of Forest Research Organisations Working Party (IUFRO) 7.02.09, Phytophthora in forests and natural ecosystems
18-25th March 2017, Sapa, Vietnam
January 2017
28th USDA Interagency Research Forum on Invasive Species
10-13 January 2017, Annapolis, Maryland, USA
October 2016
Phyto-threats biannual all project team meeting
5 October 2016, APHA, Sand Sutton, York
Improving nursery resilience against threats from Phytophthora
Phyto-threats workshop
6th October 2016, APHA, Sand Hutton, York
April 2016
Phyto-threats start-up meeting
21st April 2016, NRS, Scotland
Phyto-threats workshop November 2019
November 13th 2019 held at APHA, Sand Hutton, York
Phytophthora disease threats in UK nurseries and wider landscapes: what’s here, what’s coming and what we can do about it
PHYTO-THREATS is a collaborative research project involving seven participating institutions from across Britain and is funded by the Living With Environmental Change partnership through the Tree Health and Plant Biosecurity Initiative. It ran from April 2016 till the end of December 2019. The four main objectives of the project are to;
- Examine the distribution, diversity and community interactions of Phytophthora in UK plant nursery systems
- Provide the scientific evidence to support nursery accreditation to mitigate further spread of Phytophthora
- Identify and rank global Phytophthora risks to the UK
- Gain a greater understanding of the evolutionary pathways of Phytophthoras

Sarah Green (Forest Research), Phyto-threats project co-ordinator, welcomed everyone and gave a brief introduction to the project, reiterating the aim to address global threats from Phytophthora species and to mitigate disease through nursery best practice. She described the outcomes of the previous two stakeholder workshops (in 2016 and 2017) in terms of evolving attitudes towards risk and accreditation and outlined the objectives of this workshop, which were to;
- Share the latest science findings from the Phyto-threats project
- Provide an interactive demonstration of science outcomes and tools
- Explore how the project’s science outcomes can best be used to support the continued development of accreditation and Plant Health policy
The meeting was attended by c45 stakeholders representing nursery managers, landscape architects and garden designers, Plant Health inspectors, foresters, academics, policymakers and others. This report provides an overview of the presentations given on the project team’s research, the Plant Healthy Assurance Scheme, key outcomes from the interactive science sessions and general discussions, and next steps to ensure impact.
Research Highlights
David Cooke (James Hutton Institute) presented the results to date from the project’s work package 1, which looked at the distribution, diversity and management of Phytophthora in UK plant nursery systems. He started by thanking the managers of partner nurseries who participated in the fine scale survey for permission to sample and ran through the objectives of this work. These were to manage the risk of import and spread of Phytophthora, generate data in support of biosecurity protocols, enable earlier detection of the next threat, and analyse the diversity of Phytophthora species in relation to nursery management systems. He showed a video of zoospore release to remind everyone that managing Phytophthora is all about managing water! He also reminded everyone of the life cycle factors which make Phytophthoras so damaging, the large diversity of known species, and then ran through the history of P. ramorum on larch as an example of why we want to stop the next Phytophthora!
David then presented the nursery sampling protocols for the fine scale surveys. This involved fifteen partner nurseries which were each sampled 4-5 times over the three-year project period, and the broad scale surveys which involved the sampling of a further 118 plant nurseries as part of statutory Plant Health inspections. The partner nurseries for the fine scale survey included a range of business types from forest tree nurseries to traders of amenity horticultural plants and garden centres. He reiterated that the sampling was not random, but targeted symptomatic plants and known Phytophthora hosts as well as management practices including water sources, drainage systems and run-off ponds. He then showed some photos illustrating the type of material sampled before running through the process of metabarcoding which identifies Phytophthora species based on their unique DNA signatures and the data analyses leading to reports of Phytophthora diversity in each sample.
The project collected 2869 root samples from 163 host genera, the top 25 of which were shown in a figure and included Juniperus, Taxus, Viburnum, Pinus and Rhododendron as the most frequently sampled species. The sample analysis is a two-stage process, with the first stage being a PCR test to determine whether Phytophthora is present in the sample or not. All Phytophthora-positive samples are then progressed to the second stage which involves Illumina sequencing to determine which Phytophthora species are present. Overall, 40-50% of all nursery samples collected were positive for Phytophthora. This varied according to host genus, with Chamaecyparis, Pinus and Fagus yielding some of the highest proportions of positive samples. In terms of Phytophthora test results in relation to nursery practices, the percentage of positive samples varied across partner nurseries from 20-70%, and this reflected the plant health status and nursery management practices observed at sampling. One key objective was to feed results back to the nursery manager to help them improve practice.
David then talked through species findings, with 51 Phytophthora species identified so far with P. gonapodyides, P. cinnamomi, P. cryptogea, P. syringae and P. lacustris as the five most common species. The clade 6 taxa (such as P. gonapodyides and P. lacustris) are generally considered native and less pathogenic and are abundant in rivers in Europe. However, the other abundant species (which, in addition to the above species, also included P. cactorum, P. cambivora, P. plurivora and P. nicotianae) are common pathogens on many hosts in the nursery industry. The important quarantine species P. ramorum has only been found in eight samples to date and P. kernoviae not found at all, whereas other regulated pathogens P. lateralis and P. austrocedri have been found in 17 and 10 samples, respectively, so far.
Some species of concern include P. cinnamomi, which is adapted to warmer environmental conditions, has an exceptionally wide host range and was found to be widespread on a range of hosts particularly in nurseries in southern England. Phytophthora quercina was found in the majority of Quercus plants sampled in the survey. This species is thought to be native to Europe and implicated in root damage and progressive decline of oaks. A DNA sequence matching (tropical) clade 5 species P. agathidicida/castaneae/cocois was found in many water samples, particularly in southern nurseries. David raised whether imported components of potting media (i.e. coir) could be implicated as the source?
David talked through some new species records for the UK and showed how outputs have been reported to nurseries with anonymised reports. He then talked about management practices affecting pathogen arrival (source and health of plant material coming in, growing media, water source, mud on vehicles/boots), pathogen spread on-site (including water management and hygiene) and pathogen dispersal off-site (quality control at sale, water run-off, plant disposal), i.e. how do we translate these findings into effective management practice? He then concluded his presentation with ongoing work/challenges, such as continued development of the detection tool, final PCR testing and sequencing of remaining samples and reporting to nursery managers, followed by meetings with nursery managers, Plant Health teams and those responsible for the development of the Plant Healthy Assurance Scheme to discuss the implications of the results and how they can be translated into improved practice and policy.
Discussion
Q: Did we see a relationship between the percentage of positive samples and type of plant or management?
A: Yes, either based on observed practices at sampling or nursery type, but generally there seemed to be a good relationship with practice. Data analyses will link Phytophthora species diversity to host and management practice to identify priority hosts and practices to target in an accreditation scheme.
Q: On the issue of plant disposal, would you recommend burning?
A: Yes, if it’s possible in your area, a pathologist would recommend this, but composting is an option too.
There followed some discussion on composting protocols, with some protocols already tested and shown to work.
David warned that a ‘hospital’ discard area should be treated as a contaminated zone – ideally don’t have discard piles and certainly don’t sell the plants as discounted or give them away (similarly with used pots).
Mike Dunn and Mariella Marzano (Forest Research) presented work package 2 studies on the feasibility of an accreditation scheme, giving perspectives from different stakeholders. Mike showed a few photos of good and poor nursery practice in terms of biosecurity and spoke briefly about best practice guidelines from an Australian nursery accreditation scheme. He then went on to describe the surveys conducted by the work package 2 team exploring perspectives on accreditation from different stakeholders including nurseries, retailers, landscaping sector, local authorities and the public.
Key findings from the consumer survey of 1500 plant buying members of the public found that they obtain plants from multiple sources (garden centres, DIY stores, supermarkets, friends and nurseries) so there is a need to consider the whole supply chain, not just nurseries, when developing accreditation. When making decisions about which plants to buy and where to buy from, factors such as quality, range of products offered, and cost are key drivers. When asked about which sources they feel are riskiest in terms of pests and diseases, it was clear that they are using sources they themselves would consider to be of higher risk (e.g. non specialists such as supermarkets and DIY stores). Thus, either education or promoting accredited products on the grounds of high-quality healthy plants is required. Existing purchases of non-horticultural products (fair trade coffee, red tractor produce etc) is driven by ideals and quality. The biggest interest in accreditation comes from the biggest spenders e.g. those who spend more are more likely to travel further for accredited plant products.
Moving on to discuss perspectives from nurseries, retailers (including garden centres), landscape architects and designers, based on 253 survey responses and 33 interviews, it was found that nurseries and garden centres have similar pest and disease concerns (e.g., Xylella and Phytophthora), but the largest proportion of landscape designers rated ash dieback as their greatest concern.
Mariella ran through some of the key comments and concerns from these sectors in terms of pests and diseases, with the reputation of the business, disruption, negative impact and the practices of other plant sellers being of concern, i.e. we might be doing things properly, but others aren’t, and the risk comes from there. Mike talked about perceptions of risk reduction by introducing a suite of biosecurity practices. Nurseries and garden centres reported an expected 25% (mean value) reduction in risks posed by Xylella and Phytophthora if they were to implement best practice, however around 55% of respondents perceived no reduction in risk though best practice. Although the best practices aimed at growers would be inapplicable to landscape architects and designers, 92% of these groups said that biosecurity and pest and disease issues influence their choice of plants when preparing planting specifications.
Landscape architects and designers may also include pest and disease precautions in the specifications. For example, they might outline that particular species should be used. This appears to be happening to some degree (around half of landscape architects and designers always/often include pest and disease precautions in their specifications). However, a limitation of the role of landscape architects and designers in the pest and disease conundrum is that there’s no guarantee that the trees and plants they specify will actually be planted in the way they intended or outlined. If an accreditation scheme became established in future, landscape architects and designers may advise that plants are obtained from an accredited grower.
Going on to look at the use of twelve biosecurity practices by nurseries, only four of the practices are currently used by the majority of nurseries. Having a quarantine/holding facility, water treatment and reliance on UK suppliers were all practices perceived to be most costly, whereas boot washing, disinfecting stations and vehicle washing stations were perceived to be least costly. Owing to a lack of clear guidance, nurseries varied in terms of how they approach biosecurity practices, such as disposal of waste/sick plants. In terms of driving good practice, retailers are influential consumers and could really help in driving good practice if they demand transparency in their supply chain, including biosecurity beyond the border.
In terms of attitudes towards an accreditation scheme, there was strong agreement from both nurseries and garden centres in; a) an accreditation scheme better ensuring the quality of trees/plants sold to consumers and b) safeguarding the wider environment from the spread of pests and diseases. So, clearly, there are some perceived benefits and agreement that accreditation would fulfil some very important aims. However, there was a mixed response in terms of interest from nurseries and garden centres in joining a hypothetical scheme. Both sectors were slightly more negative than positive, but the largest response category was in the middle (50/50 whether they would join a scheme). This might be because they were being asked to consider a hypothetical scheme with no detail on how it would work and what it would cost. During the afternoon’s interactive session Mike and Mariella ran an exercise to consider which factors stakeholders think are most important for an accreditation scheme to be successful.
Mike then ran over some of the challenges to accreditation such as reasons for non-adoption of the twelve best practice criteria used previously in the survey. So, after asking nurseries about what practices they did use, they then asked why they didn’t use the others. When offered a range of response options the most common reason was ‘inappropriate for size of business model’. The perception that ‘we don’t need those practices’ is perhaps the biggest barrier, even more so than cost! Again, the importance of these practices in pest and disease prevention may not be fully appreciated. There was some cynicism among nurseries and garden centres about insufficient interest among growers for accreditation to be successful, and the costs of such a scheme for growers and consumers. Mariella talked through some of the comments received from nurseries and garden centres in relation to their concerns over accreditation.
When asked about willingness to pay for the business and the stock to become accredited relative to their existing business costs (% premium), strikingly, the largest response category for nurseries and garden centres was 0%. However, if the costs can be kept low, say 1%, then 64% of nurseries and 60% of garden centres would be prepared to pay for accreditation.
For landscape architects and garden designers, their impact is limited because the schemes they specify for aren’t always carried out as they describe. Going forward, the option to specify that landscape contractors should attain plants from accredited buyers might be possible. However, real change could require those procuring contracts to put biosecurity concerns ahead of other factors (i.e., how timely or cheaply a site can be planted). If this were the case, demand for accredited products would increase, and interest in becoming accredited or stocking accredited tree/plant products would likely increase.
Mariella and Mike finished up by summarising their findings so far in terms of appetite for accreditation;
- Accreditation must cover multiple businesses, or at least their stock
- Many nurseries will have to improve their biosecurity practices to become accredited
- Few nurseries are willing/able to incur substantial cost to become accredited
- What accreditation ‘looks’ like (teeth, costs and benefits) will be influential
- More evidence is needed of how regulated pests and diseases could impact growers, and how biosecurity practices avert such risks
- Public are driven by quality rather than biosecurity practices. Thus, quality could be emphasised to promote an accreditation scheme
- A requirement for large landscape contracts to use stock from accredited growers would increase demand (and therefore suppliers’ interest).
- Retailers are willing to work with science and policy to improve practices and could serve as an important player in raising awareness of plant health
Discussion
Comments:
Online purchasing is a problem – how does that fit into accreditation?
Retailers don’t want negative messaging. Lots more discussion going on about biosecurity and raising awareness and providing peer pressure to change behaviour.
Need to establish the Plant Healthy assurance scheme.
Virtual nurseries – a huge problem.
Need to make accreditation compulsory.
Needs to be a requirement for accreditation with logo on the door of the nursery. Xylella is making a difference in a positive and negative way. There are nurseries in Italy who are in trouble and are keen to sell so are dropping prices and this puts pressure on some to buy cheap. Oak processionary moth is another huge problem – close to 60+ sites with it. Some nurseries were careful when importing oaks, but others shipped oaks without any paperwork.
Beth Purse (Centre for Ecology and Hydrology) presented on the results of work package 3 linking global spread and impact of Phytophthoras to biological traits, trade and travel, suitable habitat and climate. She described the process of pathogens as invaders, going through the stages of transport, introduction, establishment and spread, with the potential for invasion failure at each stage in the process. Beth’s team have been looking at which traits or biological characteristics enable a Phytophthora species to become successful invaders, overcoming all the barriers along the way.
Beth showed how Phytophthoras spread by trade and travel, presented a map of trade flows into Britain, and talked through how these pathogens are sensitive to climate and habitat conditions. The main aim of her team’s work has been to analyse pathogen behaviour and traits to identify and rank global Phytophthora threats to the UK. The specific questions they have been looking at are; i) how are Phytophthoras introduced into the UK?, ii) which species arrive?, iii) which species establish, and which parts of the UK are at risk?, iv) could tourism present a potential pathway of introduction?, and v) what range of hosts and sectors in the UK could be impacted?
In order to start looking at the above questions, Beth’s team developed a global Phytophthora distribution database from a range of sources, collating close to 40,000 country-level records covering a range of sectors from garden/amenity, forest, nursery and agriculture. They also developed a database of Phytophthora traits for all 179 described species, together with collaborators in Australia and New Zealand. Beth ran through the Phytophthora biological traits considered to be important at each stage of the invasion process. These include whether the species produce resilient resting structures such as oospores, chlamydospores and hyphal swellings, their thermal tolerances for growth, whether sporangia (sacs containing infective zoospores) are caducous (i.e. ‘deciduous’ allowing aerial dissemination) and whether they cause root disease or aerial disease, or both. They hope that the trait-based framework can be used as a predictive tool, so that when a new species is first described, key morphological or biological traits can be measured as a first priority to determine potential impact.
Looking at risks of introduction, Beth showed a map of annual live plant imports into the UK since 2000 in which it was clear that most live plant imports come from countries in the EU (e.g., The Netherlands and Ireland), but also that there is significant trade in live plants from the USA. Plants are also imported from Asia and Australasia – the global network is extensive. To determine what factors most influence risk of arrival of a new species into a new country, they looked at 35 Phytophthora species with more than one documented arrival in any country since 2000 and associated trait data, taking into account national recording and biosecurity effort in each country where these species have been reported. They found that introductions were strongly correlated with the level of connectivity to source regions by the live plant trade. They also found that some species are better able to exploit trade pathways than others, and that these tended to be cold-adapted Phytophthora species. Beth then showed the model that she will demonstrate in the afternoon session where, for any country, the user can look at import volumes of live plants from source countries and the Phytophthora species present in those countries. For recently discovered Phytophthora species (i.e. post 2014) the model also provides a link to trait-based predictions for future global spread of those species.
Moving on to risk of establishment, Beth’s team have collated a very large dataset of records of Phytophthora species detections in the UK from various sources. She presented a map of the UK showing locations where P. ramorum has been detected, as an example (including nurseries and the wider environment). They found that the more common Phytophthora species were recorded in nurseries as well as in forests and gardens. The thermal traits of species appear to have a strong effect on the latitudinal range of spread with cold-tolerance in Phytophthora linked to establishment at higher latitudes.
For the question ‘how much of the UK is at risk of a particular Phytophthora species establishing?’ Beth showed some global niche models using the already well-established invasive species P. cinnamomi and P. ramorum as ‘training models’. These models are still under development, but show very nicely the riskiest UK regions where each species has indeed established. Essentially, models with environmental factors and global occurrence data give accurate predictions of distribution in the UK wider environment. Different Phytophthora species vary according to which environmental drivers are important to their establishment in a region. For several species (P. cactorum, P. cinnamomi, P. cryptogea and P. plurivora), the seasonality of precipitation was the major driver. For P. ramorum, however, winter temperature appeared to be the most important factor.
Essentially, global niche models are a valid methodology for identifying suitable habitat for Phytophthoras in the UK. However, lots of occurrence data are needed to develop the models and this is not available for many species that are yet to arrive in the UK. Also, many
Phytophthoras are unknown to science when they first emerge in the invaded range. Centralised Phytophthora occurrence and trait databases integrated across sectors and enhanced sampling in Phytophthora source regions are needed to be able to develop models and predict species behaviour when invading temperate areas.
Beth then moved on to whether biological traits explain variation in global spread and host range of Phytophthora species. They looked at geographical extent (number of countries ‘occupied’ by that species) for 156 species, and number of different plant hosts recorded for 145 Phytophthora species. They found that cold tolerance, the ability to infect roots and the ability to cause foliar symptoms were the traits most associated with geographical extent. Wider host ranges were strongly linked to optimum growth rate, oospore wall thickness (i.e. long-term survival) and ability to cause both root and foliar symptoms. They then asked whether thermal traits, especially cold tolerance, modulates invasion of Phytophthora into temperate regions. There are certainly indications that cold-tolerance may be more labile than heat tolerance which could have implications for colonising new regions, particularly those at higher latitudes. Looking across the Phytophthora phylogeny (classification) thermal traits and cold tolerance, which Beth’s team have linked to invasion success, are very variable suggesting recent adaptation.
Summing up findings and horizon scanning; connectivity to source countries through the live plant trade is strongly linked to introduction, and species that are cold-adapted are better able to exploit the live plant trade pathways. In terms of risk of establishment in the UK, cold-adapted species establish further north in the wider environment. This is predictable from global niche models, and the global climate limits of each Phytophthora species are linked to their biological traits. In terms of impact, phylogenetic relatedness is as strong a predictor of global impact as biological traits. Years known to science is also important as biological and distribution information is gathered on each species over time. Cold tolerance and ability to cause root and foliar disease predict geographic extent. Thick oospore walls, faster optimum growth rates and ability to infect both roots and foliage lead to wider host ranges. Closely related species are similar in impact. Phylogeny + traits explain 50-60% of the variation seen in host range and geographical extent for Phytophthora species, so these factors could be used in horizon scanning when looking to predict the potential impact of newly discovered species. However, the traits looked at in this study were those traits measured routinely for species descriptions – are we missing other key traits linked to invasion?
Beth concluded her presentation by stressing the importance of the global Phytophthora databases developed as part of this study being available to researchers and other end-users, being updated as new species are described and information becomes available. She also showed how her team’s model outputs can help influence policy and practice, for example the Phytophthora importation tool, models for predicting species’ likely impact in the UK, and another tool listing Phytophthora hosts in UK forests and commercial forestry, which could be used to inform species choices for commercial planting and afforestation. Beth asked the audience whether they would be interested in using any of these tools to inform their practices, and if yes, whether their practices could change as a result of the tools. She also asked what modifications or additional information would they like to see in the tools or models to improve usefulness, and how best to access the tools, i.e. website, phone app? It was hoped that useful discussion on these questions could be had during the afternoon’s interactive demonstration of the tools.
Discussion
Q: Do you have any Phytophthora data from Ireland?
A: Yes, we have data from both Northern Ireland and the Republic of Ireland.
Q: Cold-tolerance (temperature) is mentioned a lot in establishment, but what about precipitation?
A: Yes, rainfall is key e.g. evidence for autumnal moisture having an impact on P. ramorum/ P. kernoviae, but also survival during dry conditions is important.
Ewan Mollison (University of Edinburgh) gave a brief overview of the work package 4 research on predicting risk via analysis of Phytophthora genome evolution. Phytophthoras are oomycete pathogens that cause serious plant diseases and there are around 170 species currently described. More species are being discovered all the time, mainly as a result of global surveys looking in source regions of suspected high Phytophthora diversity. We know that the impact of Phytophthora varies among species and that when a new species is discovered it is very hard currently to predict its impact. One of the bases for this study was to see whether impact can be predicted by the genes that a species has through a study of Phytophthora genomics.
After an explanation of what is meant by ‘genomics’ (determining the entire DNA sequence of organisms and fine-scale genetic mapping), Ewan ran through the rationale of the work, which was to sequence the genomes of three Phytophthora species currently regarded as less-damaging, but which are closely related to highly damaging species. By comparing the genes present in less damaging species with those of highly damaging species, it might be possible to find genes present in damaging species, but absent in less damaging species that might be linked to virulence. Ultimately, gene content might help us to predict which newly discovered species are likely to have most impact.
So, the project team set about sequencing the genomes of i) P. europaea which was first described from soil associated with European oaks, but which is closely related to P. alni, a hybrid species killing riparian alder across Europe, ii) P. foliorum, currently known only to cause a minor foliage blight of azalea and rhododendron, but which is closely related to P. ramorum which is killing larch in the UK and tanoak in the USA, and iii) P. obscura, first associated with horse chestnut and pieris, but closely related to P. austrocedri, which is killing juniper in the UK and Chilean cedar in Argentina.
Ewan ran through the process of genome sequencing, which is to extract the total DNA from each organism, fragment the genome, apply a new sequencing technology to generate millions of sequence reads from the short DNA fragments, then use computer software tools to assemble the overlapping DNA sequence reads to produce a complete genome. In order to be able to look at the gene content of a genome with any accuracy, it is very important to have as complete a genome assembly as possible, i.e. to have the final genome construction in as few fragments (known as ‘scaffolds’) as possible. Using the latest sequencing and assembly technology, Ewan and the team were able to produce three of the most complete Phytophthora genomes yet produced for a Phytophthora species (at this time 30 Phytophthora species have had their genomes sequenced) i.e. the P. foliorum genome assembly has ~ 60 million DNA base pairs assembled into just 22 fragments.
Each of the three genomes sequenced in this project have ~19,500 predicted genes and Ewan presented two slides with figures showing a comparison of the genes present in each of these three less-damaging species with the genes present in closely related highly damaging Phytophthora species. The figures showed how many genes are shared and how many are unique to each group of species. Initial analyses revealed 40 genes present in highly damaging species which are not present in the less damaging species and which might be linked to virulence. The next steps are to analyse the function of these genes in order to start to unravel what makes a Phytophthora virulent. Gene content can then be used to predict which newly discovered Phytophthora species have the potential to be most damaging.
Update on the Plant Healthy Assurance Scheme
Helen Bentley-Fox and Amanda Calvert (Grown In Britain) presented an update on the Plant Healthy Assurance Scheme. Helen is the technical manager for the Grown In Britain standards, and Amanda is an auditor with nursery expertise. For the last one and a half years they have been working with the HTA and Alistair Yeomans to develop the Plant Health Management Standard which will form the basis of the Plant Healthy Assurance Scheme.
Essentially, the Plant Health Management Standard is a checklist for anyone who works with plants to deliver plant health. It is based on the International Plant Protection Convention’s framework for pest risk analysis. The idea is for plant producers to use this checklist and be subject to periodic reviews to help them improve their plant health management. The Plant Health Alliance Steering Group is the governing body for the Plant Health Assurance Scheme which provides the overarching implementation of the Plant Health Management Standard. Growers can sign up to the scheme, be audited against the standard and publicise that they are part of the scheme through use of the Plant Healthy logo. The scheme details how companies are audited, by whom, how often and how. The intention is that Plant Healthy unites all other grower accreditation schemes into a single all-encompassing scheme. Although the scheme has not yet been rolled out, an online Plant Healthy self-assessment tool is available and allows businesses to identify where they could improve their biosecurity before joining the scheme. Around 200 people have completed the online self-assessment to date.
The Plant Health Management Standard has a list of 23 requirements that demonstrate that a business is operating responsibly. These 23 requirements are designed to improve the health of the plants that are bought and sold, and to enhance biosecurity. The tricky bit with the standard was to not be over-prescriptive. They aimed for something generic enough to cover all businesses, but specific enough to be auditable and effective. The Standard is available on the Plant Healthy website https://planthealthy.org.uk/resource-topics/sector-guidance-documents with guidance documents provided for all the different sectors right down the plant supply chain from propagators to landscapers. These documents explain how each sector can apply the Standard to their businesses.
The Plant Health Management Standard was developed under two key concepts; i) Appropriate Level of Protection (ALOP; World Trade Organisation) and ii) Pest Risk Analysis (PRA; International Plant Protection Convention). These are normally reserved for nations, but have been applied here to a ‘site’ or business to enable a joined-up approach to plant health biosecurity. Amanda and Helen presented a slide showing the pest risk analyses procedure for each business as a cycle; essentially the boundaries of the ‘site’ (business) are identified, the plant species handled within that site identified, any potential pests and diseases affecting those plant species identified, the pathways of introduction of each pest and disease mapped, and the level of risk to that business if controls are not in place identified. The control measures are then identified and the degree to which each measure can mitigate the risk. The Appropriate Level of Protection is defined for that site and monitoring put in place to demonstrate the effectiveness of controls and management systems. It was emphasised that this is a continual process and not static. Underpinning the cycle of risk assessment is training of staff and recognition of risk.
The latest update on the scheme is that the Plant Health Biosecurity Alliance are meeting this month to approve the business plan, secure funding and the certification process. The scheme will be launched to all trade sectors in spring 2020 with a ‘soft’ launch to the public later in the summer. They need to train more auditors however and the Plant Healthy logo can only be used for the company, not on each plant, as the plants might move on to a business not applying the standard.
Amanda then ran through the audit process, which are covered in the following steps;
- An online form is filled out by the company to enable the auditor to plan the audit and the process, i.e. which members of staff do they want to see etc?
- The auditor arrives on site and makes observations of the surrounding environment.
- Paperwork and site assessment: staff are interviewed, information gathered on location, size, no. of employees etc. The director must be present as they are the lead and their influence is disseminated down.
- The legal requirements are then assessed; plant passports, phytosanitary certification, forest reproductive material regulations. Evidence and records are looked at followed by on the ground spot-checks.
- The business plant health policy is examined, this policy is not their detailed written records of protocols, but the higher level aims. Each site needs a statement scoped according to their business.
- Plant health responsibilities must be assigned to staff with a flow chart of people involved. Staff are interviewed to determine how information flows along the chain.
- The business plant health risk is identified, and an assessment done to ensure that the specific plans and protocols are in place to reduce these risks to an Appropriate Level of Protection – are the controls fit for purpose?
- Supply chain management; businesses must show evidence of having risk-assessed their suppliers.
- Plant health hygiene and housekeeping measures are assessed, such as the growing media and soil, water and weed management, cleaning (i.e. dirty pallets), waste treatment and disposal, site water drainage and run-off, vehicle cleaning – they ask, for example, ‘where you were last?’
- Plant health controls are checked to ensure good records and data, including all plant labels and codes. Plants coming in – where do they go to after they arrive at the site and are they checked for health? Same for plant dispatch. How are complaints managed?
- Requirement for ongoing self-assessment of plant health and for continual staff training and recognition of plant health competencies.
Discussion
Q: What if someone fails the audit?
A: They do not get the logo, but if they are close to passing then a progress plan is needed. They are given feedback and are graded so they know which areas need more work and these, especially, will be checked for progress the next year. Auditors are trained so that they all apply the same level of evaluation to each business.
Q: How does this link with other accreditation schemes?
A: The aim is for a single, overarching accreditation scheme, which is Plant Healthy.
Comment: It is hoped that the outcomes from the Phyto-threats project will feed in, and inform, this scheme.
Comment: Yes, we have a standard, but the way it is written means that novel information will enable the company to take this information on board to continually upgrade their processes.
Q: This is for nurseries. What about a garden centre? If plants from an accredited producer sat next to a sick plant from somewhere else?
A: Yes, we plan to extend the scheme to retailers to make sure the system goes right down the supply chain.
Q: How will arboriculturalists benefit? How will you get them on board?
A: The standard can be applied to arboriculturalists. Both the Landscape Institute, British Association of Landscape Industries and the Arboricultural Association have their own best practice guidance and the scheme references these.
Q: How prescriptive do you plan to be for water testing? For example, if filters need to be cleaned, do you insist a test is the best idea for a filter and insist on it being done?
A: More prescriptive conditions may come as scheme develops, but it’s not currently prescriptive. We highlight a problem area and say that it is a problem and ask how they will tackle the problem, this is an example of not being overly prescriptive – how they tackle it must suit their business and they can choose any number of ways. We would make a point of checking during the next audit. The idea also is that it will encourage businesses to improve; for example, training, if they need to update knowledge in certain areas.
Comment: The Phyto-threats project will identify a set of priority management practices for the standard, likely based around water source and usage, growing media, raising plants off the ground. These could be referenced in scheme guidance.
Interactive demonstration of project tools and outcomes
Best practice and water treatment options
David Cooke presented on nursery best practice in relation to Phytophthora findings at nurseries so far and Tim Pettitt (University of Worcester) presented different water treatment options; the pros and cons.
Outcomes: there was much discussion on what to do with the waste plants and growing media. It’s a big challenge and the options are burial, burning, composting. Drying and incineration was suggested as an option, also pelleting and burning-green energy.
Q. Is it OK to use rainwater collected into a tank? (asked twice)
A. Rainwater can become and often is contaminated with oomycete plant pathogens, surprisingly small amounts of infected debris on roofs or more likely in gutters can cause problems – often contaminated rainwater will have a wonderfully clear appearance. On the ‘up’ side, rainwater is very good quality generally and easy to treat to remove plant pathogens with any of the available technologies which all work well on this water source.
Q. On UV effectiveness with increasing turbidity of water.
A. You can pre-filter, but even cleared water may have UV-dense materials, such as dissolved organic materials that reduce the transmittance and render the treatment less effective. Also, even tiny particles (<25 µm) can scatter UV light and cause problems with treatment efficacy.
Q. How can you be sure that wetland species don’t propagate Phytophthora?
A. We can’t! However, monitoring of two commercial irrigation water treatment systems treating water with floating flag iris has consistently shown removal of Phytophthora and Pythium spp. (although not related oomycete species Saprolegnia ferax). Also, similar systems have been apparently used successfully in Zundert in the Netherlands since the early noughties. The mechanism for these ‘removals’ or population declines is not properly understood and the possibility of the iris roots in such systems becoming infected by pathogenic Phytophthora or Pythium species with wide host ranges has not been properly explored. (more information is available in AHDB review CP126, see pp 90-92)
Q. What’s the environmental footprint of waste treatment? (costs vs carbon footprint)
A. I don’t think this has yet been effectively studied in-depth. All water treatment systems will have capital costs for equipment and storage, and these vary between treatment systems, with some having lower installation costs (e.g. in-line dosing systems). Running costs will involve power for systems such as UV and ozonation and especially pasteurisation as well as increased amounts of pumping to move water through systems and to/from storage, whilst dosing systems have the recurrent cost of the chemicals used. Full assessments of carbon footprints can only really be properly achieved on a case by case basis and I don’t think this information is yet available.
Q. How often should you change MyPex matting?
A. It depends what the MyPex is sitting on and its general condition. Obviously, if it becomes damaged or heavily contaminated with mud and debris it should be replaced. However, if it is on top of a reasonably well drained gravel or sand bed, and so long as debris and weeds such as moss and liverworts are removed, then it should be possible to use sterilants like Jet 5 or similar to clean the matting and remove pathogens between crops and matting should only need replacing when it becomes physically damaged. It’s impressive how big an impact simply washing and sweeping debris off the surface of matting can have in terms of reducing pathogen inoculum, although this alone is no panacea!
Feasibility of accreditation
Mike and Mariella ran an exercise to consider which factors stakeholders think are most important for an accreditation scheme to be successful in terms of uptake and impact. Participants were presented with a list of 9 factors which may influence success, as determined through surveys and interviews with actors from across the horticultural supply chain. These factors included:
Each person was asked to rate the following options in order of importance:
- Right biosecurity practices i.e. those with a demonstrable impact on reducing the introduction and spread of pests and diseases
- Greater public awareness
- Greater industry awareness
- Buy-in from those contracting planting schemes (local authorities, conservation bodies etc.)
- Low-cost membership
- Carrots for participants e.g. assurance that they won’t be punished for the presence of pests and diseases when seeking to improve biosecurity
- Sticks for participants e.g. penalties for failing to meet a scheme’s biosecurity standards
- Scheme branding – emphasising healthy plants (quality) and safeguarding the wider environment as well as the use of a recognisable logo
- Involvement of industry in scheme’s establishment and evolution
Participants were invited to reflect on and discuss these factors in order to clarify what each encompassed. Participants were then encouraged to suggest other additional factors which may determine a scheme’s success. Once a shared understanding of the factors was reached, participants were tasked with a voting exercise designed to elicit the relative importance of the factors. Specifically, each participant allocated a finite number of points (using 10 stickers) across the list of factors, with more points reflecting greater importance. In all, 37 people participated in the exercise including representatives of landscape architects, garden designers, government, Plant Health inspectors, environmental charities, nurseries, botanic gardens, conservation volunteers, the RHS, arboriculture consultants, commercial foresters and research.
The outcome, presented in Figure 1 below, revealed that low-cost membership was considered the most important factor to ensure an accreditation scheme’s success. The ‘right’ biosecurity practices emerged as the second most important factor, followed by a number of factors relating to increased awareness of pest and disease impacts, biosecurity practices and the scheme itself. The most commonly suggested ‘other’ factor which could determine a scheme’s success was ‘mandatory involvement’.
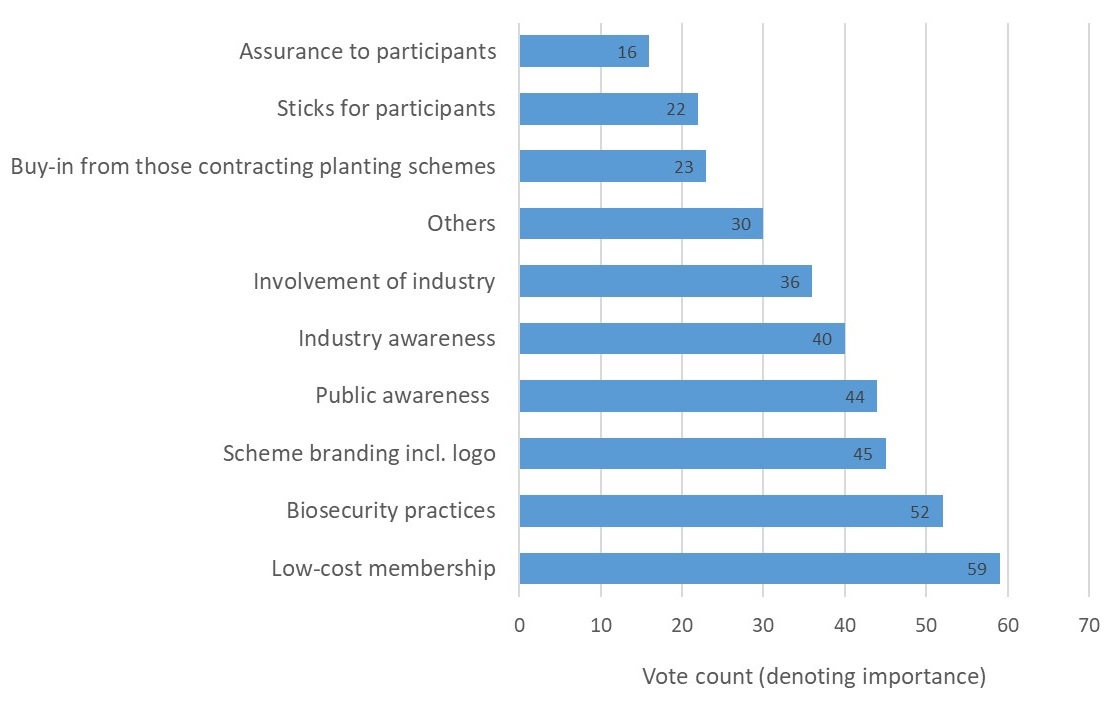
Figure 1: Factors for accreditation success as ranked for importance by 37 stakeholders.
Demonstration of modelling tools to predict risk
Beth and Louise demonstrated a collection of online tools in development, which would allow users to interact with the data and modelling outputs from the Phyto-threats project.
Stakeholders were asked
- Would you use any of these tools to inform your practices?
- If yes, how could your practices change as a result of using these tools?
- What modifications or additional information would you like to see in the tools or models to improve their usefulness?
- In which format would you like such tools to be delivered? e.g. website, phone app.
The session explored three tools with stakeholders:
- Where are Phytophthoras coming from?
This tool focusses on the role of international trade in the movement of Phytophthoras and allows users to visualise the Phytophthora diversity in exporting countries and the volume of imports from those locations. Stakeholders were asked whether this would be a useful way to assess the risks of importing plants from different origins.
Q: Could the tools include information about Phytophthora and seed imports?
A: This will depend on whether trade data identify seeds within the commodity types – Beth and Louise will check if this information is available in their trade data.
The trade data in the tool sparked a useful discussion about the lack of information about traceability:
Comment: It is useful to know specific countries that are potential sources of Phytophthora, but the trade flow data do not capture the complexity of plant supply chains, which can involve multiple steps between countries. The poor traceability of plants prior to the previous country in the supply chain was highlighted as a problem for nurseries when buying plants.
Comment: Plant passports from Defra can tell us an origin, but plant only needs to be in UK for six weeks to become British.
Comment: APHA are making efforts to trace back origin of outbreaks for particular tree hosts.
Comment: Phytophthora source data could be useful when sourcing plant material from further south in Europe, in the context of climate-proofing.
Comment: One envisaged application was to look at threats from fungicide resistance in the pathogens in the country of origin. If importers knew this, they could think about not importing plants from some countries if it’s too risky (e.g. China fumigate all plants from Europe and pressure wash, so there are opportunities to tighten up biosecurity).
2. Which Phytophthora are associated with hosts in UK forest and forestry?
This tool is an interactive database of Phytophthora pathogens and associated hosts, which is searchable by host and by pathogen and links to risk maps of UK suitability for the pathogens.
Q: Could the tools be rolled out to cover a broader range of pests and pathogens?
A: The data for a number of other pests and pathogens of concern like Emerald ash-borer and Xylella are available from the Centre for Ecology and Hydrology, where Beth and Louise are based.
3. Prevalence of Phytophthora species in UK nurseries and the wider environment.
This tool allows users to interact with maps of Phytophthora disease records in the UK. The maps show which species are predominantly nursery-associated and which are common in the wider environment. Host information is available by hovering over the record on the map.
Q: Will the tools be password protected, or will nursery locations be protected when displaying Phytophthora records? In many regions, nurseries could be identified even if the data were shown at 10 km resolution or even county.
A: Only Phytophthora reports from the wider environment (not nurseries) will be displayed when the tools are made publicly available.
General questions and comments from stakeholders:
Q: What is the added value of these tools beyond what is already in, for example the UK plant health risk register and plant portal, the EPPO alert list or the Plant Healthy portal?
A: The tools should provide more information about the relative risk of these species.
Comment: The Risk Register now has thousands of pests and diseases which makes it difficult to know what to zero in on.
Comment: Beth and Louise need to think about how it can be made relevant and linked in with industry? It would be of value to run a series of workshops to draw in key users and trial the tools.
Q: Could the tools be made simpler for risk assessment?
A: Model outputs could be translated into estimates of relative risk (e.g. ranking species or countries).
Comment: Visual nature of tool is very helpful. Perhaps it could be used to raise public awareness of threats from Phytophthoras and motivate the public to ask nurseries about what they are doing about pathogens.
Key outcomes
There was general agreement that a website would be the most useful format to access the tool. Potential uses of the tools were identified including identifying high risk Phytophthora species, hosts and source countries for users at different stages within the plant supply chain, to prioritise plant health surveillance and as an educational tool for the public. Improvements were also highlighted including simpler and more interpretable metrics of risk, broader coverage of pests and pathogens (a one-stop-shop) and the resilience and substrates used by different Phytophthora species. There was an appetite among stakeholders for further co-development of the modelling tools.
Role of genomics in predicting risk
There was considerable discussion around how genomics tools can assist future management. One of the ways to assist international plant health protocols is to be able to undertake a quick genetic test to determine whether a newly discovered species is likely to be highly virulent. We are a long way from that at the moment, but our work aims to take us there, if possible. For example, Phytophthoras have genes known as RXLR effectors which are linked to virulence. Some of these effectors may be more key than others in determining virulence. The number of these effectors varies greatly among species, e.g. about 350 each in P. sojae and P. ramorum and over 550 in P. infestans. These effectors tend to be associated with rapidly-evolving regions of the genome and may even play a role in how Phytophthoras can adapt to host resistance.
There was discussion on host jumps;
Q: P. ramorum jumped host, so how related to larch were its previous hosts?
A: Not related at all! Larch is the first known conifer host for this pathogen.
Q: What makes a species jump host, genetically? i.e. can a genome study help us to identify the potential for a species of Phytophthora to jump to a new host?
A: We don’t know this at the moment, but that is something that might become apparent as genomics advances.
Q: Is there potential for a species of Phytophthora to wipe out an entire host species?
A: Hopefully not if there is genetic diversity in the host population, where some individuals will have genetic tolerance to the pathogen. Larch is a commercial forestry species which is probably not very genetically diverse and is often grown in single species blocks, allowing rapid spread of the pathogen through the susceptible crop. With P. austrocedri on juniper, which is genetically diverse, we think we are seeing resistant individuals which do not develop disease. Hopefully their offspring will carry this resistance to allow a certain amount of population recovery.
A: We don’t know this at the moment, but that is something that might become apparent as genomics advances.
Q: Which clades of Phytophthora are most damaging?
A: There’s no real correlation between clade and damage – more a case of clades tending to be less damaging within their own “native” environment: e.g. David Cooke’s comments about clade 6 species being less of a problem in the UK.
Q: How much does copy number variation influence pathogenicity of Phytophthora?
A: Some evidence that elevated copy numbers or higher ploidy can lead to a more aggressive Phytophthora (e.g. in P. infestans).
Q: Are genomic methods used for other pathogen types?
A: Yes – widely used with other pathogens, e.g. RNA-Seq experiments to examine changes in host gene expression during infection; and genome sequencing of fungal, bacterial and viral pathogens.
Q: Has any work been done using knock-outs or knock-down experiments to investigate gene function in Phytophthora?
A: Not that I know of, but would be an excellent way to verify experimentally genes involved in pathogenicity.
Q: What about using GM to control Phytophthora?
A: Might be an option with regard to developing resistant plants (but runs into a minefield of laws and regulations!).
Q: Can we work out the age of Phytophthora species?
A: With phylogenetic analysis of multiple gene sequences we can track changes backwards to estimate at which points different species diverged.
Q: Are there similar genomic studies being used to predict potential hosts/resistant plant species?
A: There is a great deal of research into identifying / selecting resistant plant species / cultivars, but not so much (as far as I know) into identifying what makes a species more susceptible.
Q: Is there a risk of biocide resistance arising in Phytophthora?
A: Definitely – as with biocide resistance in any pathogen, there will always be some cells that are less susceptible than others and excessive or inappropriate use of biocides risks creating an environment where these have a competitive advantage.
General discussion
The workshop concluded with a short general discussion session with the following questions, answers and comments from the stakeholders and project team;
Q: Should accreditation be mandatory?
A: It makes it viable.
Comments:
A mandatory accreditation scheme means costs and who pays?
Science projects are good, but strong links are needed to be sure accreditation is science based.
Demonstration of best practice on nurseries might help to get buy-in from nurseries?
How to engage with the ‘hard to engage’, for example contractors?
Next steps
The project science team will continue to liaise with the Plant Healthy Assurance Scheme in terms of helping to develop the Plant Health Standard and in securing uptake and consumer support. We will also liaise with policymakers and practitioners over predictive models to ensure that our project outcomes are used to support pest risk analyses and the risk register.
Phyto-threats project team meeting November 2019
November 14th 2019 held at APHA, Sand Hutton, York
The aim of this meeting was to bring the entire project team together to share and discuss research progress since the last all-project team meeting on November 20th 2018, to outline planned outputs and to discuss next steps for continuing collaborations now that the project is finishing.
The science updates for each work package were presented the day before at the stakeholder workshop and a full report on each work package is available in the stakeholder meeting report published online:
Therefore, the report here focuses on additional information not presented at the stakeholder meeting and on the resulting discussion.
WP1 Phytophthora distribution, diversity and management in UK nursery systems – David Cooke (JHI) and Peter Cock (JHI)
David Cooke started off by emphasising the need to get back to the different organisations with an interest in the outcomes of this work package and to think about how best to engage. He planned to show today the Phytophthora species findings by nursery and for Peter Cock to show progress on the bioinformatics pipeline. They also need to link Phytophthora findings to nursery management structure with Beth’s team.
While David was running through the slides a question was asked regarding clades;
Q: Are you going to give a breakdown of clade when reporting back to nursery managers?
A: We had decided not to report clade, especially as sometimes clades change.
Comment: Nursery managers don’t need to know clade.
Q: But, isn’t there a clade-specific treatment or approach? i.e. clade 6 species tend to be less damaging than clade 8 species.
A: Grouping by clade might be risky, for example sending out a message that all clade 6 species are ubiquitous in water courses and not problematic. What about P. pinifolia for example? That one is a damaging needle pathogen of radiata pine in Chile.
Comment: We have this unknown clade 5 sequence cropping up in water samples at several nurseries. Clade 5 are regarded as tropical species, so this is most likely to represent a tropical Phytophthora and we think about it differently. Basically, if it affects how a species is dealt with then we should report clade.
Comment: Should we then include information on traits for each species found? i.e. provide a link in each nursery manager’s report to the traits’ database online. This means we put a simplified version of the traits’ database on to the Phyto-threats website and in our report to nursery managers we say ‘For more information on each species found and clade, follow this link…..’ It would have to be digestible for general use so should say e.g. oospores, chlamydospores, soilborne, airborne, damaging to which host etc.
Comment: Clade 11 has to be included somehow.
FR are currently pulling together distribution maps based on UK findings of Phytophthora species and it was suggested that these maps also link in to the Phytophthora traits’ database when that goes online. This would be useful as it would allow nursery managers to see the distribution of those species in the UK.
Comment: The species distribution maps should differentiate records from cultures vs DNA findings. There is a danger of reporting a species based on DNA findings only when Statutory Plant Health policy is currently based on the need for a viable culture.
Comment: We need to look at technology and how it relates to policy – does Plant Health policy need to catch up with technology and start to base actions on DNA findings too?
Comment: The sequences of all the species will go on GenBank; a few species can’t be differentiated using ITS1.
Q: Ultimately, could you offer this metabarcoding technology as a service for the public good? i.e. as a testing service?
A: That could be possible. This metabarcoding method would really lend itself to being developed and used as part of statutory plant health surveillance of nurseries. There are already some commercial companies specialising in eDNA barcoding, so this could be an extension of the services they offer. Water in nurseries could be tested, and then re-tested every few years. This might be a tool to assist in the issue of phytosanitary certificates.
Moving on with the presentation, David showed a slide listing host genus sampled by survey type, i.e. fine scale and broad scale, to illustrate the different range of hosts sampled. In the broad scale survey, the hosts were largely ornamentals with viburnum the most commonly sampled host, followed by rhododendron, camellia and pieris. David then showed the percentage of Phytophthora-positive samples for the broad scale and fine scale surveys – all around 40-50% positive.
Q: Can we relate broad scale survey results to practice?
A: We only asked for 5-10 samples per broad scale nursery – with each sample taken from different batches of plants. On average we received 6.3 samples per broad scale nursery which is possibly too few to link to practice. Also, associated metadata were not collected for the broad scale survey, only host symptoms description. However, APHA and SASA have knowledge of the background of each nursery, for example import history might be useful – a plant may have arrived with a Phytophthora and not acquired it in that nursery. We have scanned copies of each broad scale sampling sheet and those can be referred to when interpreting results.
Back to the presentation, David showed that over 70% of DNA sequence reads produced so far have matches to known Phytophthora species on the database. However, 7% of reads match unclassified Phytophthora species. So, it appears that there are quite a few unknown species in the samples.
Q: What about the ypt gene which has been used as an alternative barcode for Phytophthora?
A: There have been some issues with the ypt gene mainly due to it being only single copy. We are planning to look at other barcoding options as a separate project.
David then went on to show Phytophthora species diversity by nursery and this generated much discussion on how management practice affects diversity. There were clear examples of how certain species predominate at certain nurseries, with nursery size, practice (in particular importation of stock) and geographical location appearing to have an effect on species diversity. E.g. P. cryptogea (broad host range) was top, followed by P. gonapodyides, P. cinnamomi and P. plurivora. Records of P. pseudocryptogea and P. cryptogea are put together. One nursery has lots of P. cryptogea. Another has almost no Phytophthoras and operates with very tight biosecurity.
Comment: The Phytophthora species that we find on trees as part of our FR advisory service do seem to match species found in nurseries. For example, P. plurivora is our most common species on trees and P. cryptogea/pseudocryptogea is very common in agricultural soils and we find it causing serious root rot of trees planted on former agricultural land. We should try to match nursery findings to FR’s Tree Health Diagnostic Advisory Service findings in trees.
David moved on to discuss ongoing work/challenges, which, in addition to the need to complete sample processing, report to nursery managers and liaise with Plant Healthy Assurance Scheme over results, included continued refinement of the detection tool – the very high sensitivity of metabarcoding is both a blessing and a curse! For example, the need to prevent and account for field and lab contamination through the use of synthetic control sequences. Also, the computational pipeline development, coping with reads beyond 1-2bp thresholds, species boundaries and visualising data outputs.
David concluded his presentation with final thoughts/conclusions; metabarcoding is a very powerful, targeted method for exploring microbial diversity. A classifier has been developed, although results are interpreted with caution. When confident, the data will go to GenBank. With expanded primer sets we could look at other oomycetes. The sample bank of eDNA offers huge potential and experiments are now needed to advance biology and ecology.
Peter Cock presented on the bioinformatics pipeline THAPBI-PICT now published online https://pypi.org/project/thapbi-pict/. He talked about technical variation in metabarcoding, determined through the use of four synthetic DNA control sequences alongside real samples. Illumina produces 1000s of sequence variants generated in very low abundance. These 1bp variants are most likely PCR artefact as single base change by PCR can happen and we need to account for that in metabarcoding diagnostics. Lots of these variants are coming through at low level. These variants were used to set a minimum threshold, which was 100. This means that any unique sequence needs to be present at least 100 times before going through the pipeline; i.e. any unique sequence at abundance of less than 100 is dumped early on in the pipeline. Sequence thresholds are then set for each plate for reporting species in a sample based on the level of contamination of synthetic control sequences in environmental samples. So, this is a plate-by-plate threshold set above the highest level of known sequence contamination.
Peter ran through how sequence data are prepared, by quality trimming FASTQ reads, merging the overlapping FASTQ reads into single sequences, discarding reads without both primers, converting reads into a non-redundant FASTA file and then filtering reads with hidden Markov models of ITS1 and synthetic control sequences so that non-matching sequences are discarded. This results in big data reduction!
Peter then showed edit graphs to illustrate the number of ITS1 variants observed for each species. Many at low sample abundance are likely to be PCR variants, but those occurring at higher sample abundance and with higher read numbers could be true ITS1 variants within the genomes, for example in P. austrocedri and P. nicotianae. There are also lots of Peronosporas and Phytopythium. Complex ITS1 clusters with up to 3 bp edits are observed for P. rubi and P. cambivora (the latter which is likely to be a hybrid complex?), but the most complex cluster occurs with P. gonapodyides, P. megasperma, P. chlamydospora, P. lacustris. All of these species are hard to distinguish by ITS1. So, the cut-off very much depends upon the species, sometimes 3bp difference works, for others we have to use just one bp difference.
In terms of interpreting sequence space, there seem to be some novel species in our sequence data. Species classification starts with a 100% match to a database sequence. If there is no perfect match, then the classifier looks for a 1bp difference. The database has ~170 individual sequences curated to species (by David) plus a much larger set of sequences from the NCBI database. Any sequences not matching the curated database, but which match the NCBI database based on 2bp difference, are reported to genus only. If it doesn’t match anything then it is marked as ‘unknown’. The pipeline does not use Swarm any more as it is too ‘fuzzy’. Looking at complex sequence clusters, these could be curated manually if we want to know what the non-matching species or genera are.
Comment: Sometimes we only manage short reads for an organism so it can’t be identified, but we have isolated it and we can see it’s a Phytophthora. The point is that we can still miss things using metabarcoding methodology.
Peter showed the outputs produced by the software tool THAPBI-PICT, for example the Excel spreadsheet of results. He has tested the classifier on other Phytophthora datasets as well as a nematode ITS1 dataset and found that it works well. The team plans to publish a paper on the THAPBI-PICT software, another on the use of synthetic controls, the nursery sampling dataset and an environmental monitoring paper. They hope to try and culture some of the novel Phytophthora species that the data hints at.
There was some discussion of the method and also on the Phytophthora community analyses planned for the nursery data. It was decided that there are enough data now to start planning these analyses, i.e. categorising factors to include in the analyses. This needs to link in with the nursery surveys that Mike and Mariella did, i.e. to categorise nurseries based on management practice/behaviour. Mariella has lots of information on partner nursery type and practice and if Beth gave her a list of what classification criteria they want, Mariella will pull it out and pass it on. The challenge with management factors is that some are easy to categorise, but others are qualitative and hard to rate, for example level of biosecurity practice. David is to give Mike a framework of what he needs, and they will provide it. Beth said she was planning to provide details on the information she needs after Xmas.
WP2 Feasibility analyses and development of ‘best practice’ criteria – Mariella Marzano (FR) and Gregory Valatin (FR)
Gregory Valatin presented on the costs and benefits of implementing nursery best practice from a nursery perspective, based on data pooled from 75 nurseries. The data come from three sources; an initial FR survey of nine nurseries, joint visits by FR and Fera to eleven nurseries and the work package 2 nursery and garden centre survey of 55 nurseries. Nursery responses were based on the perceived costs and benefits of 12 best practices (water testing for pathogens, water storage in fully enclosed tanks, water treatment facility, clean/covered storage of growing media, installation of drains or free draining gravel beds, raised benches, disinfectant stations for tools, quarantine holding area for imported plants, composting/incineration facility for diseased/unwanted plants, boot washing station, car washing station, buy only from trusted or accredited UK suppliers).
One main issue with collecting such data are the high number of non-responses when each nursery was asked to estimate the costs of installing each best practice. For most of the best practices, the majority of nurseries did not provide an estimate. In addition, the numerical responses that had been provided were dominated by a few high estimates, resulting in a skewed distribution of cost estimates. Quite a lot of the nurseries are already using some of the best practice measures, so the analysis needed to consider a baseline of what measures are already being implemented. The assumed baseline of practice included five of the most commonly used best practices – those currently used by the majority of the nurseries indicating whether they used a specific practice, or not. These were; water storage in fully enclosed tanks, clean/covered storage of growing media, installation of drains or free-draining gravel beds, raised benches and tool disinfestation stations. For the purposes of comparing the costs of establishing and maintaining the best practices with the anticipated benefits, a ten-year time horizon was selected, reflecting both the length of time before many of the best practice infrastructure would need to be replaced and the investment decision time horizon used in practice by the one nursery for which this information was available. Nursery respondents were also asked to estimate the costs to their business of a Phytophthora outbreak and (in the case of the nursery and garden centre survey) of a Xylella outbreak.
Gregory asked how many Phytophthora outbreaks could nurseries avoid by adopting best practice? This created some discussion as generally it is very hard to predict with any accuracy how many disease outbreaks can be avoided.
Q: Isn’t it better to take a measure of the price benefit of being part of accreditation, i.e. by adopting best practice the nursery can charge more for its plants than other nurseries?
A: But, if every nursery adopts best practice and joins an accreditation scheme then (apart from that associated with increased plant health/quality) there is no price advantage! It’s better therefore to stick with the data we’ve got (also given there was no time for further survey work).
Gregory suggested that if reduction of risk of an outbreak is too complex to consider at the moment (perhaps it requires a new project to address this, by looking at nursery characteristics and environmental factors and modelling the frequency of outbreaks) then perhaps we need to think in terms of how many outbreaks you avoid during a certain period until the benefits outweigh the costs? He then presented an exploratory cost-benefit analysis based on the implementation costs for the seven measures which are not common best practice currently and the perceived benefits (avoided cost) of avoiding outbreaks. Based upon mean costs and benefits, this indicated that the costs outweighed the benefits where avoiding Phytophthora outbreaks alone is considered and there are one or fewer avoided outbreaks a year over a ten-year period. As these are widely considered relatively infrequent, the analysis suggested that the costs outweigh the benefits of implementing best practice based on avoiding Phytophthora outbreaks alone. (The costs of the best practices amounted to roughly nine times the costs saved by avoiding one outbreak). However, if the potential benefit of avoiding an outbreak of Xylella was considered in the analysis, then the benefits of nursery best practice outweighed the costs if one outbreak of Xylella were avoided over an eight-year period, or one outbreak of both Phytophthora and of Xylella avoided every 9 years. These estimates were based on using the mean costs and benefits from the survey data, and assuming a 3.5% discount rate (standard in government appraisals). Considering instead median costs and benefits – which are arguably more typical of nurseries given the skewed distribution of estimates, suggested for the benefits to outweigh the costs, there would need to be at least one avoided Phytophthora outbreak every 4 years, or an outbreak of Xylella avoided every 7 years, or one Phytophthora outbreak and one outbreak of Xylella avoided every 11 years.
A further factor that could also usefully be considered is the impact best practices have on reducing risks of other pests and diseases. However, no estimates were available in the survey data for these benefits. This analysis focuses upon the perspective of nursery costs and benefits, without accounting for the costs of outbreaks in the wider environment, for example, in forest and amenity landscapes (which is the focus of modelling work by Colin Price).
There was some discussion on the costs of disease outbreaks. Nursery managers expect to lose some plants each year. What threshold of disease/mortality causes nursery managers to take action? It might be that an ‘all or nothing’ loss due to an outbreak is unrealistic and that a general erosion of loss (quantity/quality) might occur over a longer time until it gets critical.
Gregory explained that he considered this analysis to be exploratory, as risk reduction is dependent on measures implemented by the whole plant trade sector and not just nurseries. It was also difficult to get enough quantitative responses from nursery managers on costs of implementing best practice and costs of avoiding outbreaks of Phytophthora and Xylella to be confident in the analysis. Some of the responses to the outbreak questions were qualitative. Gregory concluded by emphasising the nurseries’ divergent perspectives on best practice and accreditation; some nurseries want stricter regulation and others prefer voluntary measures, particularly ‘traders’ who import many of their plants. Some nurseries adopting best practice consider other plant traders to be exposing them to unacceptable risks – an aspect that from an economic perspective could be characterised as an ‘externality’ (uncompensated effect of activities over which they have no control by another company).
For those nurseries already investing in accreditation, questions arise as to how they perceive risks and pass on accreditation costs.
Discussion then focused on frequency of disease outbreaks in UK nurseries and how to quantify this? We could look at interception data and look at the number of positives found at different nurseries for each pathogen. A future project needs to pull together all the different data to get a good bottom line. Gregory’s study has found just how hard it is to get good data in order to estimate costs.
There are also risks of disease outbreaks imposed on society, and the societal perspective will be provided by scenario analysis using Colin Price’s CARBBROD model. Colin was too unwell to attend the meeting, but he put together a PowerPoint presentation available here on the economic costs of tree diseases, with the predominant impact being on carbon fluxes. Some of the presentation is a recap of that presented at the 2018 Phyto-threats project team meeting, and some of it an update. Colin was looking for feedback on a number of inputs for the CARBBROD model which will be used for a cost-benefit analysis of introducing nursery best practice from a societal perspective, in which the carbon impacts of tree disease epidemics are considered key. Gregory asked the questions for Colin, which ranged around the likely reduction in risk of a hypothetical Phytophthora outbreak by introducing best practice, the percentage of total UK area of Sitka spruce and oak susceptible to introduced Phytophthora, and the likely rate of spread of Phytophthora in both hosts. There was much discussion of these questions, the first one being particularly hard to estimate! Some estimates were pulled together finally for Gregory to pass on to Colin, with further discussion to be had by email.
It was suggested that Colin could find useful a forestry model for felling larch for P. ramorum control. Jane can help with this. Beth and Louise also have references to forward to Gregory.
Mariella gave a summing up of the work package 2 research which looked at the feasibility of accreditation through exploring the perspectives of a range of consumers. Over the course of the project they have surveyed or interviewed 75 nurseries, 61 garden centres, 183 landscape architects or designers, 18 landscaping contractors, 15 local authorities and 1500 members of the plant buying public. They are now busy coding all the qualitative data for analysis and working on improving landscape architect and contractor data. This is being done as part of the BRIGIT project on Xylella. Progress this year includes publication of two technical reports on the consumer and nursery/garden centre/landscaper/surveys which have been very popular. A journal paper on the public consumer survey has also been published in ‘Plants, People, Planet’. Future publications will include a collaborative article on the role of landscape architects in mitigating the spread of plant pests and diseases, another paper on the sector appetite for accreditation – opportunities and challenges (with Tim Pettitt) plus other papers on the role of tourism in spreading plant pathogens, biosecurity practices and the cost-benefit of biosecurity measures led by Gregory.
Mariella asked what other outputs should be produced this year? They have loads of data, where could it be used? The outcomes need to go out to growers, but how do we do this? How can it change their behaviours? It was suggested that Mariella’s team produce two sided fliers outlining key results for producers and which could go out to Plant Health inspectors. Their results should link to the Plant Healthy team. Articles in trade magazines are another good way of reaching out to the industry. Possibly soundbites on changes introduced by participants already e.g. I’ve made small changes at not much cost by doing this and this is the benefit.
WP3 Global Phytophthora risks to the UK – Louise Barwell (CEH)
Louise Barwell focused her presentation on global niche models being developed by Dan Chapman as they are looking for feedback. She showed examples of the global niche models being developed using P. cinnamomi. Their aim was to train the models using distribution data from outside the UK and then use the models to predict suitability within the UK. The approach is then validated by using distribution data from within the UK. For P. cinnamomi, there is an excellent match between predicted and actual distributions. Importantly, the UK data were not used to train the model so that a completely independent dataset was used to validate the predictions.
Dan was able to fit models for a further nine Phytophthora species with sufficient distribution data (more than 75 records from outside the UK) to make a niche model. All of these species are in the UK so are useful for validating the methods to be used for species not yet in the UK. The main issue here though is that for most species not yet recorded in the UK there are insufficient records from outside the UK to produce a model.
Louise listed the drivers which the project team thought would be important in risk of establishment – plant host distribution came out top, followed by pathogen traits (spore/inoculum density). She then showed some correlations between environmental temperature and Phytophthora species’ cardinal temperatures for growth. The correlations are good, showing that species tend to be temperature-constrained. Accounting for the invasion process used a model developed by Dan and co-authors in a paper published earlier this year (J Biogeography 2019 46 1029-40). They looked at Phytophthora known occurrences and defined a 100 km buffer around known presences based on a literature review of Phytophthora dispersal distances conducted by a summer student, Katy Roy. Model performance is good for non-UK Phytophthora in terms of discriminating presences and absences.
Louise showed the visual outputs for global niche models for P. ramorum and P. cinnamomi, and then presented a figure showing the model outputs for relative importance of different environmental drivers affecting risk of establishment among Phytophthora species. For all species the climate variables seemed to be more important that land cover, although this should be broken down by forest type to better reflect the hosts. For P. cambivora, P. gonapodyides, P. lacustris, P. ramorum and P. alni the minimum winter and mean summer temperatures are the most important drivers, but for P. cactorum, P. cinnamomi, P. cryptogea and P. plurivora, the more important driver is precipitation seasonality. Based on these drivers the model poorly predicted the UK distribution of P. plurivora, but gave a good prediction for P. ramorum. Louise asked whether the project team thought the drivers being used in the model were realistic? For example, is there a more appropriate predictor of establishment than precipitation seasonality for P. plurivora?
Comments:
Prior to 2009, P. plurivora was known as P. citricola, which might explain the poor prediction. The findings of P. plurivora seem to be concentrated around south/south-east of England and the Midlands, but we are finding it more frequently in Scotland over recent years. There have also been outbreaks of P. plurivora on lime trees in Cornwall, which don’t appear as locations on the model. The most updated distribution data for this species needs to be shared. It is possible that pathogen introductions and outbreaks are mediated by human activity so do not necessarily correlate with climate suitability.
True that for P. cinnamomi the seasonal precipitation is very important. It was only possible to bait for P. cinnamomi at a particular site in Spain following rainfall.
P. ramorum outbreaks followed when there was a lot of summer rainfall. So, spring 2013 and 2018 had outbreaks after wet summers in 2012 and 2017.
David will send Louise distribution maps for P. infestans;
The P. ramorum prediction fits well with where a species is present. Louise was directed to look at data from Northern Ireland for her distribution.
It was pointed out that sometimes climate preferences don’t accurately map occurrences, for example chestnut blight which was found outside predicted areas because it was introduced to those areas.
Louise then asked where the Phytophthora traits and global occurrence databases should be hosted. There was some discussion about having them hosted on the new IDphy tool which is replacing the Phytophthoradb website. However, Beth felt we should try to look for further funds to maintain the database on our Phyto-threats website and try to include intra-species variability. Beth also thought that as part of a grant proposal we could look at bringing the Phytophthora community together to link the different Phytophthora databases. It was decided that Beth’s team should contact the collaborators in Australia and New Zealand and discuss how to take the traits and occurrence databases forwards and where/who to host them.
Louise updated everyone on papers with two currently in submission on the traits-based analyses of Phytophthora impact; more are planned, including one on trade pathways, one on the niche models based on UK data, i.e. what makes a species able to establish outside nurseries? Other outputs include a list of Phytophthoras which affect current and future forestry species. Ana offered to send Beth a list of all Phytophthoras the FR THDAS has found infecting plantation forestry species in the UK.
Beth and Louise finished up by demonstrating the tools and model outputs showing global trade pathways and risk of importation. They have 35 species with presence in exporter databases and source countries based on data since 2000. There was lots of discussion around how the tool could be improved and how to broaden its relevance to a range of stakeholders. It was suggested that the model is circulated to the project team to have a play with. It needs coding for each Phytophthora/host record to rank in terms of reliability of the record, with isolation of the species as a viable culture better than PCR + sequencing which is better than metabarcoding.
A suggestion was made that the tool could be used to explore what pathogen might be coming, with which host it is associated, and an estimate of risk i.e. if you increase your trade in this area the risk might increase to this! Some practices have risk to the environment. It would be useful for every sector, but the big players moving large quantities of plants should come on board. It could be a useful learning tool.
Q: Based on which criteria does a species make it into the model?
A: We looked at data since 2000. If there was a reference, source and distribution to connect it was added. So, there are 35 species.
Q: Does the model assume free trade?
A: It is based on actual observed trade patterns.
Q: Can you model the risk if the supplier is ‘UK-grown’ only.
A: The problem is that trade flows may not reflect the real pathway. Netherlands (NL) to UK may be Italy to NL to UK. And ‘homegrown’ labels are applied when an import has been in a UK nursery for as little as 6 weeks in some cases.
Q: Can you interpret data to show closely related species behave similarly?
A: Yes, some do, but some behave differently! Relatedness is only part of the story.
Comment: Nursery data will be removed before making it accessible to all because currently it may be possible to zoom in to an area and correlate an identification with a nursery.
WP4 Predicting risk via analysis of Phytophthora genome evolution – Ewan Mollison (University of Edinburgh)
Ewan Mollison updated the team on the genome sequencing of three Phytophthora species currently regarded as less damaging, but closely related to highly damaging species. These were; i) P. europaea, a clade 7 species which was first described from soil associated with European oaks, but which is closely related to P. alni, a hybrid species killing riparian alder across Europe, ii) P. foliorum, a clade 8 species currently known only to cause a minor foliage blight of azalea and rhododendron, but which is closely related to P. ramorum which is killing larch in the UK and tanoak in the USA, and iii) P. obscura, another clade 8 species first associated with horse chestnut and pieris, but closely related to P. austrocedri which is killing juniper in the UK and Chilean cedar in Argentina. The rationale behind the work is that by comparing genes present in closely related damaging and less-damaging species we can identify genes or gene families involved in Phytophthora virulence.
Ewan ran through the genome assembly process; the genomes were sequenced at the University of Exeter Sequencing Centre using PacBio and two ‘SMRT’ cells to ensure high genome coverage. The assembly was done using ‘CANU’ with a 100 Mbp estimate. Ewan presented a table with the assembly statistics, essentially showing high N50 values and a low number of contigs for all three genomes. The contigs were scaffolded with ‘SSPACE-Longread’ and gaps filled with ‘gapfinisher’ over several rounds, then run through ‘Arrow’ through three rounds of polishing. The resulting assembly statistics looked pretty good, with genome sizes of 77 Mbp, 61 Mbp and 62 Mbp for P. europaea, P. foliorum and P. obscura, respectively, assembled in 39, 71 and 29 scaffolds, respectively. However, the larger number of scaffolds for P. foliorum raised concern and these were checked further using ‘blobtools’ to identify the taxonomic origin of the scaffolds in the assembly. In P. foliorum they found 49 very short scaffolds with a lower GC content than the rest of the assembly; in P. obscura there was one such scaffold and none in P. europaea. It was determined that these scaffolds were likely bacterial contamination (firmicutes) and they were manually removed from the assemblies.
The final assembly statistics are very good indeed, with the number of scaffolds for P. europaea, P. foliorum and P. obscura reduced to just 39, 22 and 28, respectively. Gene prediction was done using ‘Augustus’ and training sets based on the closest available relative; the three species were found to have ~19,500 predicted genes. Looking at the N50 and scaffold counts across all 30 available Phytophthora genomes, it is clear that the three species assembled in this project have the most complete Phytophthora genome assemblies to date, with the closest published assembly being P. sojae. Looking at completeness of coverage using ‘BUSCO’, which provides a measure of genome completeness based on an analysis of near-universal single copy orthologs, it is clear again that our three genomes have among the lowest levels of missing orthologs, being rated by ‘BUSCO’ as 98-99% complete.
To compare gene complement, Ewan used protein sets from 26 Phytophthora genomes assessed as greater than 85% complete using ‘BUSCO’. From ~55,000 total gene clusters containing sequences from any of the 26 species, a core set of ~8,700 clusters common to at least 75% (20 or more) of the genomes was identified and, of these, over 5,000 can be classed as single-copy orthologue clusters, containing a single gene from each of the species represented by that cluster (i.e. no paralogues or duplicates, etc.). Looking at gene content differences for clade 7, Ewan compared genes from damaging woody-host infecting species and non-woody host-infecting species together with the less-damaging P. europaea. His analyses pulled out 101 genes present in the other selected pathogens, but not in P. europaea. Further assessment found that 36 of these genes are in fact present in the P. europaea genome, but are disrupted by stop codons or indels that might affect expression or function.
Looking at gene content differences in clade 8, Ewan selected three aggressive pathogens that can all infect woody hosts (P. cryptogea, P. ramorum and P. austrocedri) and compared gene complement in these species with the sequences of less-aggressive P. foliorum and P. obscura. Downstream filtering found 40 genes common to all three damaging pathogens, but not present in P. foliorum or P. obscura. The next steps are to dig deeper into the presence/absence gene sets and assess the putative functions of genes found to be absent in the less-damaging species. In terms of papers planned, they are drafting one paper on the assemblies of P. europaea, P. foliorum and P. obscura and comparative genome analyses, and a second paper on the P. austrocedri genome and isolate resequencing analyses.
Ewan received a number of questions following his talk;
Q: Are you seeing telomeres – or centromeres – in the genome data?
A: Yes, we are seeing some kind of repeats that look like a signature of this.
Q: How good are your rDNA assemblies? Can you determine rDNA copy number? If so, this could be added to the genome papers.
A: I can send you the ITS data for these three species, so we can look at rDNA assembly in relation to copy number.
Q: Has P. foliorum been found again in the UK? – so far it has only been found in Scotland on a rhododendron leaf. However, inoculation experiments have found it to cause lesions on larch.
Comment: We are picking up DNA signatures of P. foliorum in our rainfall and wind vane traps in Scotland so it might be more widely distributed than we think. So far, we have not had any findings of P. foliorum causing significant damage to any host through natural infections.
WP5 Synthesis and integration – Sarah Green (FR)
Sarah Green rounded off the work package sessions by giving a brief overview of project management, i.e. three board meetings held over the last year and minutes posted on Huddle, the last all-project team meeting report written and posted on the Phyto-threats website. In terms of stakeholder engagement members of the project team have attended the National Plant Show and Four Oaks show, also various scientific conferences including two IUFRO meetings and a Phytophthora symposium in New Zealand where Mariella and Sarah did a double act presenting on Phyto-threats work. Researchfish will open in Feb/March again for outputs. Sarah wasn’t sure when a final project report is due and will get in touch with Debbie Harding of BBSRC to ask this. In terms of publications, Sarah is still planning to co-ordinate the writing of a project overview paper for which she has a draft available, will keep plugging away at this, perhaps circulating for inputs in the new year. David Cooke also promised to put out an interim report on the broad scale sampling for SASA and APHA.
Discussion of future options to continue collaboration – Sarah Green (FR)
Various leads were discussed as potential future work areas, for example following up on the growing media as bark-based (peat-free) growing composts are not always heat-treated. There is also the issue of whether anything is coming in on coir. These components could be tested for Phytophthora.
Sarah mentioned a proposed EUPHRESCO topic that she is coordinating on early detection of Phytophthora in EU nurseries and traded plants. The idea is to roll out the nursery testing and metabarcoding protocols to EU and third countries. So far eight different countries have expressed interest in the topic. Discussions are ongoing. However, the funding levels are not high and might only be £25K (if Defra commits).
Assessing different metabarcoding tools (i.e. barcodes) for Phytophthora would be very useful and David thought there might be some JHI seed corn funding for this – December application, usually 10-20K.
Beth mentioned a NERC call ‘Constructing the Digital Environment’ which might cover our activities collating data on Phytophthora outbreaks and pathogen behaviour. Beth will look more closely at the call and what it means. Deadline is January.
David mentioned the upcoming eDNA conference, there might be funding there for environmental monitoring.
Mariella thought there could be mileage in exploring funding for evaluating impacts of science and novel information on biosecurity-related behavioural changes and how it influences policy.
NERC/BBRSC might also have money to look at interactions of Phytophthora, in particular at whether species compete, whether they hybridise: NERC treescapes call is coming! This is more ecological so more about restoration, provenances, climate change, woodland expansion etc.
Following the end of this discussion the meeting was closed and Sarah thanked everyone for their contributions to what has been a very enjoyable and productive collaboration.
List of Participants
| Contact | Organisation |
| Jane Barbrook | Animal and Plant Health Agency |
| Louise Barwell | NERC Centre for Ecology & Hydrology |
| Peter Cock | James Hutton Institute |
| David Cooke | James Hutton Institute |
| Mike Dunn | Forest Research |
| Debbie Frederickson Matika | Forest Research |
| Sarah Green | Forest Research |
| Kelvin Hughes | Animal and Plant Health Agency |
| Mariella Marzano | Forest Research |
| Ewan Mollison | University of Edinburgh |
| Ana Perez-Sierra | Forest Research |
| Beth Purse | NERC Centre for Ecology & Hydrology |
| Alexandra Schlenzig | SASA |
| Tim Pettitt | University of Worcester |
| Gregory Valatin | Forest Research |
Phyto-threats project team meeting November 2018
November 20th 2018 held at Forest Research Northern Research Station, Roslin
The aim of this meeting was to bring the entire project team and members of the Expert Advisory Panel together to share and discuss research progress since the last all-project team meeting on April 23rd 2018, and to outline and receive feedback on research plans including proposed funding for continuing strands of the project beyond this financial year.
WP1 Phytophthora distribution, diversity and management in UK nursery systems – David Cooke and Leighton Prichard (JHI)
David Cooke started off his presentation by introducing a new member of the WP team; JHI bioinformaticist Peter Cock replaces Pete Thorpe who has taken up a position at St Andrews University.
Nursery samples
To date the team have completed 59 sampling missions across the 15 partner nurseries, collecting 4016 samples of which 2165 have been PCR tested for Phytophthora. The lab team have determined that processing the buffer for DNA, rather than the filter on which the DNA was originally collected, gives lots of DNA yield, so they are no longer processing filters. The team are still working their way through the fine-scale nursery root samples.
The broad-scale sampling has resulted in root samples from 101 nurseries; 36 in Scotland and 65 in England/Wales. With 5-10 root samples per nursery a total of 653 samples have been returned. Top hosts returned from the broad scale sampling are: Rhododendron, Viburnum, Pieris, Juniperus, Chamaecyparis, Hebe, Fagus, Olea, Taxus, Prunus. It will be interesting to see which hosts turn out to be Phytophthora positive. These root extractions are under way. For the OPAL wider environment sampling of water courses (now finished) a total of 26 samples have been received.
Sample processing
Much effort has gone into root extraction optimisation. JHI tried automation with a Qiagen kit, but the result was not good. They are automating with PowerPlant Pro kit, which is currently being used for the fine-scale root samples.
PCR test results to date show that ~ 50% of samples are positive overall. The frequency of positives varies from nursery to nursery (ranges from 30-70%). These results will be presented by management practice, not by nursery, in public fora. Positives for roots are slightly higher than 50%. There are 1600 samples still to process.
Since April this year more samples have been sequenced using Ilumina on plates containing nursery samples as well as samples from other Phytophthora metabarcoding projects – which all help to validate the results. David gave an overview of the Illumina rationale which Leighton will cover in his talk later, reminding everyone that sequencing outputs are 250 bp reads, 15M barcode reads, 156K reads per sample. Synthetic controls are included on each plate to give an estimate of technical error rate and sensitivity range. Also included on one of the plates were 45 samples from cultures of known Phytophthora spp. David went on to present some of the nursery sample findings from two of the nurseries:
- megasperma /P. gonapodyides clade 6
- cryptogea clade 8a
- obscura clade 8d
- quercina clade 12
Plus some (closely related) downy mildew species
Samples from another nursery yielded DNA reads matching:
- nicotianae clade 1 from Choisia sp
- occultans/P. citrophthora/ P. terminalis clade 2 from Pachysandra sp,
Data on Phytophthora species detected in nurseries will feed into the community modelling (linking with WP3) which will assess factors such as nursery size, management practices, suppliers and geographical location on Phytophthora species assemblages detected. Nursery Phytophthora data will also be linked to Phytophthora data from natural ecosystems in concurrent projects (i.e. Ponte) and community eDNA /metabarcoding.
Next steps are to complete sample processing and barcoding, re-run samples where contamination is suspected, validate the data with the new bioinformatics classifier (involving Leighton and Peter Cock), and feedback data to nursery managers and the WP3 modelling team.
Discussion
During the discussion a point was raised that some PCR positives initially reported back to nurseries as Phytophthora positive have subsequently been identified by sequencing as downy mildew species i.e. other oomycetes. How we are reporting back to nurseries in light of this?
It was agreed that downy mildews are also important pathogens and should be reported to nursery managers, but not included in the data as Phytophthora positives.
Another comment related to the urgent need to report back to nurseries soon as they are overdue sample results, also, that it would be interesting to see if there are any changed behaviours according to results. David agreed, and said we would be able to relate photos of incidence/symptoms to results in the feedback.
It was asked whether peat or peat alternatives had been tested? The answer was no, potting mix was not tested as part of this project.
A final question: can the species data be related back to circumstances/ best practices? The answer was yes, this is the plan for the analyses.
Leighton Pritchard (JHI)
Leighton’s presentation focused on what was really being measured by Illumina sequencing. He reminded us that there are real stakes from our results (i.e. the consequences of a regulated pathogen turning up in a nursery might mean closure) so we need to be held to a higher standard of accuracy than other types of studies.
ITS1 marker sequences are used in the metabarcoding analyses. We assume one species means one ITS1 sequence, but this is not the case!
Leighton ran through the metabarcoding method and described three ‘pinch points’. These are:
- Having a comprehensive database
- Whether barcodes are precise enough
- And how ‘individualising’ are the oomycetes ITS1 sequences?
Synthetic control sequences are used to help set an error rate for each sequenced plate and to check for levels of cross contamination in test samples. Synthetic control mixes are comprised of 4 unique sequences with a base composition similar to Phytophthora ITS1. In the tests that Leighton conducted (on one of the plates) the four synthetic sequences were used at six different combinations and at three different dilutions i.e. 18 synthetic control samples in total on the plate.
Analyses of the Illumina output showed that some synthetic control samples contain 1000s of sequence variants, differing from the actual control sequences by up to 10-15 bp. How much is real variation? Leighton presented a plot showing the variants and how different they are from the original sequence that was put in. Leighton also spoke about the dilution effect: at the most dilute, worryingly, an artefact of 1 snp difference is seen. Some sequences can’t be matched to the input sequences and are likely to be contamination. These occur at a threshold of 100. So, setting a threshold above 100 gets rid of a lot of variants. If a DADA2 clean-up is performed, the threshold is reduced to 20 variants. Also seen are PCR artefacts in low abundance (i.e. when PCR amplification introduces variation) and what appear to be contaminating sequences from environmental samples. So what is in the contamination? Many look like ITS1 sequences; many don’t. Most cross-contamination comes from soil bacteria, some are oomycete sequences. The synthetic sequences pop up too at low abundance in some of the environmental samples. These are easily recognised as cross-contamination and their abundance (in terms of read counts) in samples across the plate can be used to set thresholds.
Leighton then went on to explore the ‘one species, one ITS1’ concept. Many species have more than one ITS1 sequence since the ITS region can occur in a Phytophthora genome in anything from 40-170 copies. If a single isolate is Illumina sequenced then one major cluster should result. One of the plates contained ~40 single isolate samples to test for ITS1 sequence variation across a range of species. However, from a single isolate up to 2000 sequences resulted! Thus a single isolate does not give 1 single sequence. The resulting clustering of sequences by Swarm produced up to 150 OTUs (operational taxonomic units) (mostly 40-60) from a single isolate. However, many of these sequences are variants occurring at low abundance. Removing all amplicons occurring at abundance of less than 20 before clustering reduces the number of OTUs per isolate to around 6.
The next question is ‘how are the OTUs present within each isolate ‘individualising’? By looking at the Jaccard distance between OTU members among isolates it can be seen that several OTUs are unique to individual Phytophthora species and some are common to several sequenced species. If one species has a specific ‘classifier’ (i.e. unique OTU) as well we can ignore the ambiguous ones.
Leighton asked ‘what is a classifier?’ Associating input sequences with species classes, this is the classifier. So if you change anything in the method, the classifier changes. Leighton and bioinformaticist Peter Cock are currently developing the automation of the ‘classifier’ process. This includes building the reference database framework based on trusted sources. The next step is to create a training dataset. This will be combinations of reads from single isolate controls, individualising sets for a species, applying and evaluating the current pipeline and applying validated classifiers to old and new samples.
Discussion
A question was asked about the timeframe by which we can have confidence in the latest plates to present the results back to nursery managers. Leighton said he couldn’t give a definite date, but hopefully fast! He thought early next year was achievable.
Another question related to whether there were data on isolations to go with these sequence outputs? It was confirmed that there were P. lateralis/P. cambivora isolations with DNA samples from the same material progressing down the pipeline. However isolations were never intended to be done as part of this project. It was mentioned that a Scottish Government-funded project is generating companion data from live (baited) organisms as well as metabarcode data from the same soil and water samples. We’re assuming that what is in a sample represents a threat, but this may not be so. Detectable live presence may be a better marker.
Another comment related to the sequencing process: the synthetic control in most abundance gave the same reads when replicated and those sequences that were spiked appeared in the correct order. The lowest dilution of synthetic sequences was not found or was equal to background noise, which is good. It shows this as a robust and accurate tool. In this respect, testing with control sequences, we seem to be at the forefront of metabarcoding.
It was pointed out that the nested PCR itself could potentially introduce error and there was general concurrence on this point given that the technique is so sensitive. It is recommended when starting up to use blanks and do a control plate before anything else so that an estimate of cross-contamination can be made. However, overall, when considering what species have been found in nursery samples to date, the results make sense. Pathogens appear on expected hosts (i.e. P. austrocedri on juniper, P. lateralis on Chamaecyparis, P. pseudostugae on Douglas fir, P. occultans on Buxus). If a result looks erroneous, this has usually been present at very low read number and is most likely to be cross-contamination. Hence the need to set a read threshold below which results are not considered as true. On the question of doing some qPCR validation of results, the answer was yes, this had been done, but as part of other metabarcoding projects.
WP2 Feasibility analyses and development of ‘best practice’ criteria – Mariella Marzano (FR), Mike Dunn (FR), Gregory Valatin (FR), Glyn Jones (Fera) and Colin Price (external consultant)
Mariella opened her presentation with a brief reminder of the objectives of WP2. She then went on to update the team as to where they are with the different sectors:
Nursery interviews – the team have conducted 19 nursery interviews so far and are aiming to have interviews completed by Jan/Feb 2019. One of the problems has been that there are so many questionnaires in circulation because of the competing accreditation schemes that nurseries have been deluged by questions and become fatigued! The team have, however, managed to interview several online retailers and garden centres, where previously they were short of inputs from these sectors. A survey company has been recruited to take up the task from here.
Landscapers – Mariella explained that after engaging with landscapers e.g. LI (Landscape Institute) and BALI (British Association of Landscape Industries) she was made aware that her team’s questions were too generic and not specific enough. This will need further discussion in the focus groups. ‘Landscapers’ are more accurately landscape architects, contractors or garden designers, each with different roles and implications for biosecurity. Furthermore, individual contracts determine how much impact landscapers have in choosing plants: they may be told what to plant; may be able to offer choices or alternatives; and depending on the client base they may or may not have biosecurity awareness. They may have access to a reference guide (for example the LI is releasing a ‘biosecurity toolkit’ document for their members). A good question to investigate is at what stage in the process is biosecurity important, if at any?
Glyn Jones and Barbara Agstner of Fera have done a lot of groundwork on cost-sharing. This will be presented by Glyn later.
Retailers/Garden Centres – Mariella attended a recent workshop that involved large retailers. Discussions indicated the feeling that customers trust that this sector is doing things correctly and expect quality, and retailers are wary of negative messaging. However, customers are now starting to ask more questions e.g. on plant origins. Through interviews one issue raised was that customers can take their diseased plants to the garden centre to ask experts for advice on ailing plants!, thus, potentially disseminating pathogens into the garden centres. There seems to be an opportunity here for a plant health message to the public.
Mariella then posed a question to the floor – the WP2 team had proposed to do a series of focus groups in the later stages of the project, so what should the focus groups be about? This will be discussed later during the WP5 presentation.
Mike Dunn – Mike has been exploring biosecurity issues with public parks/gardens. Both sectors have an interest in plant health standards, which is driven by the obligation they feel to meet visitor requirements, not driven by pest/disease concerns. Generally, visitors want and expect to see exotic species. The National Trust has bronze/ silver /gold plant health standard checklists of what factors to consider when procuring. Mike and team have also developed a Local Authority question framework and plan to interview 15-20 Local Authorities.
The consumer survey, including gathering of more economic data, is now going to be the responsibility of a survey company and telephone surveys will be conducted on 50 each of nurseries, garden centres and landscapers, to be completed by February 2019. Questions will elucidate location within the supply chain and key biosecurity factors.
Discussion
Mariella asked for input on how to integrate all the data from the surveys of the three main sectors and how to disseminate advice. It was agreed that packaging up information and advice from the project and making project data accessible to stakeholders would be key.
A comment was made that the team’s choice of target groups was correct, particularly the landscapers. The ‘landscaper’ sector could have some biosecurity weaknesses, because following the planning and approval stages there are apparently few subsequent checks and balances to ensure that plans are followed.
Glyn Jones presented on the Defra Future-Proofing Plant Health (FPPH) work of relevance to the Phyto-threats project, outlining three projects as follows;
Early warning system/pathways analysis – Glyn presented a slide outlining the structure of the project, incorporating figures on global imports /exports, global tariffs, gathered from governmental data and the World Bank, used to look for trade anomalies using algorithms/machine-learning. This highlighted non-usual datasets.
Industry data – this project is revealing the difficulties of actually obtaining species import data. For example one company provided 240 combinations of species and specifications. The work has highlighted the very complex network of business connections that changes from quarter to quarter across the year. They are looking at certain species and asking; how much is brought in, and what is the UK demand? For example, lavender, for which the demand is high, so could there be a good case to increase production internally?
Costs and responsibility – much of the work has been done by Barbara Agstner (Fera) as part of her PhD. Barbara has been on a road trip around Britain visiting various nurseries and getting estimates as to the costs of implementing various biosecurity measures. This work has again highlighted the many gaps where costs are unknown. One company was able to provide costing for a range of activities – this is being shared with Gregory.
In the second part of his presentation, Glyn presented on the emergence of various industry accreditation initiatives over time, i.e. BOPP, Grown in Britain, UK Sourced and Grown, the Plant Health Alliance. Currently the HTA, Grown in Britain and others have been working on a Plant Health Management standard and assurance scheme which is ready for launch in early 2019.
Any scheme will be about applying a plant health management standard(s) that will be owned by the governing body. The standard would then be adopted by the various assurance schemes. Individuals apply to the relevant certification body to join their scheme – thus currently there are several competing schemes all of which have to adhere to the agreed standard.
A scheme will have self-assessment questions, a full audit checklist, will require an audit. Everyone would sign up to the scheme and all would have to meet the criteria. However, certification schemes could compete – they would just apply standards, and would set their charge.
Discussion
The question was asked as to what best practice recommendations are needed to feed in to the standard from Phyto-threats? i.e. step-wise introductions of recommendations into a common industry standard since certain infrastructural changes need to be applied, such as water storage and treatment, raised benches, drainage, etc to make a scheme work at preventing disease. The idea of competing schemes did not seem very efficient either and stakeholders had preferred a single, overarching UK scheme at last year’s stakeholder workshop.
Gregory Valatin spoke about the cost-benefit analysis of introducing best practice in nurseries from a nursery perspective. Following an initial low response from nursery managers to the economics questions included in the WP2 survey questionnaire, Gregory joined Barbara Agstner on some nursery visits as part of the costs and responsibility sharing Fera project. Over the summer 2018 a FR intern also helped gather economic data.
Gregory listed 12 ‘best practices’ which formed the basis of the questions. Nursery managers were asked for estimates of the cost of implementing these practices. The best practices included water testing, water treatment, storage of water in fully enclosed tanks, clean/covered storage of growing media, use of raised benches, disinfestation stations for tools/containers etc, boot washing station, vehicle washing station, quarantine holding area for imported plants, installation of drainage systems, composting/incineration system for disposal of waste plants and buying only from trusted/accredited suppliers.
In addition to questions about the costs of implementing the best practices, growers were also asked for an estimate of the cost to their nursery were it to be affected by a future Phytophthora outbreak. The cost of implementing the best practices can then be compared with the expected avoided cost of the outbreaks expected to be prevented by implementing the best practice measures. The benefit of preventing outbreaks partly depends on the frequency of the outbreaks expected to be avoided through introducing the best practice measures. It is anticipated that other members of the Phyto-threats team – especially those working on WP1, may be able to advise on the frequency of outbreaks prevented (with this information also needed for wider exploratory cost-benefit analysis from a societal perspective).
Gregory presented preliminary conclusions for a number of scenarios based on economic data gathered so far (for which there are many gaps). These included where a nursery implements all 12 best practices (i.e. the baseline is that none of the measures have been implemented to date), as well as a scenario where a nursery implements just 8 of the best practices (assuming a ‘common practice’ baseline of 4 measures implemented to date). In each case it was assumed that at most one Phytophthora outbreak per year would be prevented. For the scenario of implementing all 12 measures, the initial survey responses indicated that the mean cost to nursery managers of implementing best practice, whether start up or where just the annual cost is considered, is much higher than the expected benefit of avoiding the costs of dealing with an actual outbreak. For the scenario of implementing the 8 best practice measures, the initial estimates similarly indicated that the mean set-up cost of implementing the best practice is much higher than the expected benefit of avoiding the costs of dealing with an actual outbreak, with the annual costs being of a similar magnitude to the expected benefit of avoiding an outbreak. For a ‘typical’ (1 ha) nursery, estimates from a sector expert also indicate that set-up costs for the best practice measures exceed the benefit of avoiding an outbreak (whether introducing all 12 best practices or the 8 best practices that initial responses to the survey indicate are not already common practice). Whether costs exceed the benefits to the nursery depends not only on the frequency of outbreaks that the nursery expects to avoid (which the responses to date suggest are currently far fewer than one per year – but potentially might increase above one per year if detection rates increase due to technological improvements), but also on any sales price premium nurseries are able to obtain once they introduce the best practices. Based on the preliminary data, Gregory questioned the level of take-up of a voluntary accreditation scheme if the perceived costs to the nurseries of introducing the best practices outweigh the perceived benefits.
Some of the qualitative responses from nursery managers support the conclusion that implementing best practice to reduce the risks of Phytophthora outbreaks is not seen to be cost-effective from a nursery perspective. However, as one of the nurseries contacted had noted, the cost of an outbreak will depend on which Phytophthora species is causing the damage. Gregory also asked what happens if we factor in the benefits of avoiding other pathogens such as Xylella? Although implementing nursery best practices to reduce the risk of spread of phytophthoras may not seem a priority for many nurseries, current biosecurity measures across the plant trade sector are viewed as inadequate by some nurseries. Indeed, some nurseries advocate urgent action by the government in shaping which practices are permitted – especially in the context of risks of introduction of Xylella, with mandatory restrictions needed that apply more widely to the plant trade than just the nursery sector. Information on costs is still being incorporated from personal interviews and telephone interviews carried out this summer. More economic data will also be gathered by the survey company over the next few months.
Discussion
It was asked whether outbreak costs were based on cost of loss of stock alone as it was pointed out that outbreak costs should include Plant Health inspector time, litigation costs, loss to suppliers etc. Gregory noted that the survey asked both about the anticipated loss of nursery stock, and other costs to the nursery, but that nurseries are frequently very uncertain about the level of costs likely to be incurred – as illustrated by the wide range of estimates and proportion of respondents unable to provide estimates for the costs of implementing specific measures. Phytophthora risks are often perceived by nurseries as low compared to other pathogens, so there appears relatively limited interest currently in implementing measures specifically aimed at reducing risks of Phytophthora outbreaks.
The comment was made that since improved biosecurity will be good for preventing all diseases, we need to emphasise this, in addition to talking about Phytophthora. Xylella should be included since quarantine holding areas and careful plant sourcing reduces the risks of a Xylella outbreak. It was reiterated by others present that core best practices work for all, and that best practice reduced the weeding/husbandry bill. Gregory noted that a question had recently been added to the questionnaire to enquire about the costs anticipated were there to be a future Xylella outbreak at the nursery, although initial responses indicated that it was also proving very difficult for nurseries to answer this.
Colin Price presented ‘some economic costs of Phytophthora: nightmare scenarios’ using the CARBBROD model– a model being used as the core element for the exploratory cost–benefit analysis of introducing best practices from a wider societal perspective. Thinking in a forestry context, disease outbreaks matter economically because of curtailing rotations and timber yields, wasting forest expenditures, taking land out of production, and by reducing carbon sequestration and storage. From a societal perspective, carbon impacts are key, owing to the high social values placed on carbon in relation to climate change mitigation. One of the scenarios presented using the model was of a new and unknown Phytophthora species entering the UK (e.g. via the nursery trade) and spreading to infect Sitka spruce plantations (based on a crop of yield class 14, age 30) along similar lines to the recent impact of P. ramorum on larch, with bare land following (if no other species replaced the Sitka on upland, acid sites); and another if Sitka spruce is replaced by noble fir. Owing to the trajectory of the social value of carbon used by the UK government for policy appraisal, infection of the Sitka followed by replacement with noble fir was found to give a better return to society than not having an outbreak! This was because of the increase in the discounted social value of carbon over time (with a lower price of carbon applying to the emissions associated with losses of the Sitka than to subsequent carbon sequestration by the noble fir). Everything depends on prices and timing! In prior discussions, Gregory had suggested potential for adopting other approaches to valuing carbon (e.g. assuming a fixed carbon value in real terms) to avoid the perverse conclusion that introducing new pests and diseases to the UK could be beneficial to society: Colin noted that implementing other approaches to valuing carbon was easy to do within the model, yielding the expected conclusion, that outbreaks modelled as above did indeed have adverse economic consequences. In worst cases, the cost through infection could be many tens of thousands of pounds per hectare.
Colin also looked at a scenario involving the speculative consequences of infection of oak stands with a new Phytophthora to the UK, assuming 30% mortality, 30% unaffected and 40% having reduced growth increment. In this case the stand increment recovers and regeneration comes from surviving trees. Forest managers can thin and choose subsequently to replant either with oak or something else (e.g. sycamore). In contrast to the scenarios for Sitka, a key difference here is that continuing to grow the same species on the same site is an option.
The CARBBROD model can also be applied to a scenario involving a new disease coming in and infecting urban trees, for example a broad host-range pathogen that can take out 10% of all urban trees. The resource baseline proposed for this scenario was the urban forest from the London i-Tree Eco project survey. Colin compared the costs of trees dying due to a new disease versus those of dying due to age. Costs in terms of the associated reduction in ecosystem services were evaluated, including loss of pollution abatement, loss of aesthetic services, and loss of carbon sequestration, as well as replacement costs’ being brought forwards. The costs need to be evaluated by species, age and life expectancy.
Colin closed his session by asking the floor for likely scenarios for a new Phytophthora coming to the UK and infecting Sitka, oak etc. For example how much of the crop is likely to be affected, what is it likely to be replaced with? Discussions with Colin over lunch and tea did not prove as helpful as hoped in trying to answer some of these questions. Wider enquiries will be instituted.
Appendix
Attention could be drawn to some reminders on CARBBROD, compiled by Colin and presented by Mariella to the mid-year project meeting. CARBBROD does not deal specifically with the economics of nursery practices, but with the effects of these practices on forestry plantations and woodland health based upon the likelihood of the outbreak and spread of a new tree pathogen (e.g. entering the country via the plant trade). It evaluates the consequences for timber production and carbon fixing of disease arising under different scenarios of tree age, consequences and management responses. In this project the model will be applied to important threatened species and genera (e.g. Sitka, oak, urban trees). It is flexible to allow adoption of different forms of carbon pricing, discount schedules and interactions.
In terms of future developments of CARBBROD, as the carbon effects are dominant, incorporating the effects of the new disease on litter and soil carbon (if these are known) would be useful, as well as adapting urban tree carbon for effects of disease, including landscape scale effects of disease if feasible and incorporating a simple application to scenarios of disease spread (as has been done previously for Dothistroma).
WP3 Global Phytophthora risks to the UK – Beth Purse (CEH), Dan Chapman (CEH) and Mike Dunn (FR)
Beth Purse presented the objectives and components of the work package which are;
Pathways of risk of introductions
Traits modulating introduction risk
Social factors and environmental/ecological traits influencing spread once here
Environmental niche models to map areas at most risk
Dan Chapman presented on risk of introduction of pathogens which they are addressing by modelling the risks based on transport networks and pathogen traits. Louise Barwell has accumulated a database of country level occurrences which includes 17,371 country level records with dates and 1,417 species x country combinations (the source of this information is the recipient country/ year of first arrival/invasion status).
A preliminary analysis of this database raised the question: ‘Can live plant trade network explain arrivals?’ There are data for total import volumes, network connectivity (imports of focal species from source countries), arrivals-trade-species-country. Arrivals detected post 2000 were from 94 countries and 56 Phytophthora species with ≥ 1 documented arrival. The result was that connectivity (total live plant imports from countries in which Phytophthora species occur) explains about 21% of the variation in new arrivals better than total imports (i.e. from anywhere).
Moving on to risk of establishment and spread, Beth ran through the original aim of the global niche models: to predict the area of extent of impact on UK tree species of global Phytophthora species that have not yet arrived in the UK (30) and 24 species that have arrived. Of the global Phytophthora occurrence data gathered, regions climatically similar to the UK give 11,497 records comprising 82 Phytophthora species in 38 countries. Data are still to come from countries such as New Zealand, Australia, South Africa and Canada.
Looking at the habitats in which Phytophthora species are recorded, most records are drawn from closed forest types, also urban areas and cropland. Many of the species for which sufficient information is available to make niche models on risk of establishment and spread are already in the UK. Focal species not yet present in the UK have very few occurrence records worldwide. So the aims of the niche models need to be revised. For those species already present in the UK, niche models will look at their potential distributions here. For high impact invaders such as P. cinnamomi and P. ramorum new global niche models will be developed with a range of environmental factors.
Records of pathogen interceptions/escapes in the UK have been gathered from key sources (THDAS, RHS, SASA, eDOMERO). Some data e.g. THDAS have related data on interceptions to subsequent spread in the different settings, linking back to traits and trade/recreation proxies. Geographical occurrence will be linked to environmental characteristics in order to predict potential distribution.
Beth then updated everyone on the Phytophthora traits’ database which was merged last year with a database held by Scion (New Zealand) and which will be maintained there. Beth finished up by outlining future work which will be to submit papers on the trait database and phylogenetic analyses and traits and global impact (by January 2019), trade models, finalise global niche models, policy briefs and co-development of final model outputs with policymakers and practitioners. An example of the latter might be the production of interactive source maps from trade and tourism models.
Discussion
A comment was made that host ranges did not seem to be included in the traits’ database, possibly because information only comes in when disease outbreaks occur. Yet host range data are very important. Generally, when a new species is found it is tested for pathogenicity on hosts closely related to the affected host, or on hosts that closely related Phytophthora species infect. It was agreed however that phytophthoras can surprise, for example host range tests were performed on trees expected to be P. ramorum hosts based on analogy with the US, but no one expected larch! It was asked if closely related Phytophthora species have similar impact? If you look at host range at the Phytophthora species level, rather than related group level, some species have a wide host range, others narrow. It was cautioned however that wide host range doesn’t necessarily mean they are more devastating on a host.
Mike Dunn presented on his work looking at tourism and recreation as a pathway for pathogens, carried out through literature search, survey of Plant Health researchers around the world, and visitor data to UK parks and gardens. Generally, it is considered true that recreation and tourism have acted as a pathway for spread of plant diseases previously, but that this pathway is perceived to be of lower risk in terms of spread of pathogens than other pathways such as traded plants and plant material.
In terms of visitor data, the 2011 Visit Britain survey on passengers visiting Britain has been useful in providing number of visitors, origin and timing of visits and, based on a questionnaire, the estimated number that are planning to visit a public park/garden while in the UK. Mike has also approached 23 parks and gardens asking for data on visitors, with a mean of 471,000 visitors pa. Very few of these sites collect data on visitor origin. Moving on to discuss domestic spread within the UK, a survey monitoring engagement with the natural environment conducted in 2017-2018 found that 62% of adults living in England reported making visits to the natural environment at least once a week.
The next steps for this piece of work are to write up the data and think about whether incoming tourists are a viable means of spread of pathogens.
Discussion
It was suggested that people travelling between (for example) National Trust properties in a single day could potentially be transferring pathogens from site to site. Bus tours, train tours etc go from site to site and it might be interesting to look at the tours and what their schedules are. Another suggestion was to check international plant propagators conference proceedings as examples of people going overseas and bringing back plant samples independently.
WP4 Predicting risk via analysis of Phytophthora genome evolution – Paul Sharp and Ewan Mollison (University of Edinburgh)
Paul Sharp explained that this work package focuses on understanding what can drive the evolution of a pathogen and allow pathogens to adapt to evolving host defences, expand host range and increase virulence. Comparative genomics may enable us to identify the genetic basis for some of the Phytophthora traits, for example evolving to infect woody hosts. Previously his group successfully completed similar work with 64 strains (genomes) of P. syringae: 38 from woody hosts and the rest from non-woody hosts. They were able to associate specific genes with woody hosts and they will try to accomplish this with Phytophthora.
The aims of this work package are to compare genes from available sequenced Phytophthora genomes, identify a core set of Phytophthora genes common to all species, identify species-specific genes or variation, sequence the genomes of three less damaging species which are closely related to highly damaging species (the topic of this talk) and study target genes/gene families known to be important for virulence.
Currently, they have Phytophthora genome information available from various sources, covering 27 species and 10 clades, but most genomes are from species in clades 7 and 8. Paul showed a phylogeny of all Phytophthora species with available genome data.
The project has recently sequenced the genomes of three less damaging species. These are;
- europaea, clade 7, originally associated with rhizosphere of oak forests in Europe and most closely related to P. alni, P. cambivora (woody hosts) and P. fragariae, P. rubi (soft fruit hosts).
- foliorum, clade 8, originally isolated from azaleas in US and closely related to P. ramorum.
- obscura, clade 8, originally associated with horse chestnut soils and azaleas and closely related to P. austrocedri.
Carolyn Riddell (FR) successfully extracted high quality genomic DNA from all three species for PacBio long-read sequencing and Ewan has been assembling the genomes.
Ewan Mollison ran through the results from the initial genome assemblies. The reason for choosing PacBio over Illumina is that it gives much longer read lengths (10s of Kbps), giving better resolution of repetitive regions and greater overall contiguity. Error is random rather than systematic, so if there is very high read coverage across the genome the errors can be corrected rather than amplifying bias.
Ewan ran through an earlier assembly of the P. austrocedri genome. This species was sequenced using a ‘hybrid’ method combining both PacBio and Illumina reads. The hybrid assembly was hampered by not having sufficient read depth of either sequence type for optimal assembly. Pete Thorpe (formerly of JHI) re-did the assembly based on the PacBio reads only, using the Illumina reads only for error correction. The outcome was a much improved assembly (862 scaffolds compared with 43,700 with the hybrid assembly). Therefore a PacBio only assembly was pursued with the three Phytophthora species targeted here using two SMRT cells to achieve plenty of reads across the genomes.
The data for the three new genomes (P. europaea, P. foliorum, P. obscura) were presented. A good overall read length was achieved for all three species across both SMRT cells and Ewan explained how he made his scaffold assemblies and gene model predictions. In terms of genome size the estimates for each species are; P. europaea 95Mbp (has more repetitive genome content); P. foliorum 70Mbp; P. obscura 63Mbp; therefore it is safe to use 100Mbp as an estimate of genome size for all three species. The number of contigs for the three species is low (103 to 127) indicating a very high degree of contiguity in all three assemblies.
Scaffolding links contigs together with gaps of known length padded out with ‘N’ characters. The number of scaffolds across the three genomes is also low, ranging from 67-77, again indicating a high quality of assembly. All three species sequenced here have a very low number of scaffolds compared with other sequenced Phytophthora species (except for P. sojae which has a very complete assembly), for example most species have over 2000 scaffolds, with P. cambivora having 120,000 scaffolds indicating a highly fragmented assembly.
The three assemblies reported here have a high completeness of assembly (98%), a low level of genome duplication (~1%) and good resolution of haplotypes. The likelihood of polyploidy is low. Looking across the other genomes the trend is for larger genomes to have larger repeat content. Looking at predicted proteins the trend is less clear. The number of predicted genes is generally similar for many of the Phytophthora genomes. P. cambivora came out as having a particularly high number of predicted proteins, but rather than being extra genes this is likely to be gene fragments incorrectly identified as separate genes/proteins.
Ewan showed an example of xylanases as a sample gene family which may be of interest in this study. Xylanases are an important class of plant-cell-wall-degrading enzymes.
There are four major xylanases which have been identified in Phytophthora; xyn1, xyn2, xyn3 and xyn4. Sequences from all xyn genes found in 30 Phytophthora genomes were aligned, and phylogenetic trees constructed for each gene. Clades 9/10 only have 2 xyn genes, clade 8 species have 3 xyn genes and clades 1-7 have all 4 xyn genes. Sequence differences in xyn occur in some species, which results in their falling outside the expected clade groupings for that particular gene.
Discussion
A question was asked about the use of the tool BUSCO for assessing genome assembly. Ewan explained that BUSCO tends to evaluate core genes, for example those in DNA replication and protein metabolism that are more ubiquitous. Checking that they are there would certainly provide a measure of confidence.
A non-biologist within the group asked for a digest on progress for a non-biologist! Much of the language used in this presentation was incomprehensible to those unfamiliar with the field. It was explained by Paul that, put simply, the three sequenced genomes have good sequences, so they can now look for genes to see if there’s an association between having a gene and having a certain trait. Also, for genes with target activity e.g. genes known to be involved in pathogenesis like xyn genes, they are now in a position to compare among the pathogens and non-pathogens.
WP5 Synthesis and integration – Sarah Green (FR)
Sarah Green presented an overview of coordination/communication events since the last meeting. This included two board meetings (June and August), the minutes of which were posted on Huddle. The April team meeting report and research updates for 2017/18 were also posted on the project website.
The date and location of the next all project team meeting were discussed. It was agreed that a meeting would be held in York in October 2019, together with a stakeholder workshop. It was pointed out that this should not clash with the Phytophthora IUFRO meeting in Sardina over 17-25th October.
Sarah, Tim Pettitt and Jane Barbrook also represented the Phyto-threats project at the HTA National Plant Show in June 2018 following the offer of a stand for free! Sarah took the WP2 consumer survey leaflet to hand out, as well as the two-sided flyer on best practice, based on outcomes from nursery sampling to date. Both were well received. Sarah conducted interviews for, and completed, 7-8 garden centre questionnaires, gaining an appreciation for the difficulties of social science research! She also presented a talk at the show, following which the ‘Gardening Which’ magazine approached to say they would stop recommending purchasing ‘bargain’ (i.e. unhealthy-looking) discount plants to their readers!
David and Sarah went to the Oomycete workshop in Boston in July 2018. Their presentations covered this project, in particular how we engaged nursery stakeholders. David said their approach with validation got praise. David, Leighton and Sarah will all present talks at the ‘DNA Working group meeting’ in Derby, 26-27 November 2018.
Discussion
Discussion followed on the nature of the focus groups that need to be held. What should they be about? How would it link to outcomes on accreditation? We need to hold them before the end of December 2019. Sarah suggested a focus group aimed at building in more with the HTA initiative and associated groups. Mariella clarified that a focus group meant an intense discussion for a couple of hours with just 8-10 people and she said that Fera should be included.
Beth suggested a group to discuss risk model outputs, perhaps toward the autumn, either within the October stakeholder meeting or as a stand-alone. At the stakeholder workshop next year Sarah would like stakeholders to receive a clear message from each WP including practical outcomes.
Mariella returned to questions that might be asked at a focus group with nurseries. For example how do nurseries react to what their consumers think? She wondered if there were any outstanding questions not covered or not covered adequately so far. If so, Mariella would welcome feedback.
A comment was made that we all use specialised vocabulary in our presentations. What words are washing over everyone else? It would be helpful not to use too much jargon for the public audience. It was agreed that we should always try to deliver the key messages to stakeholders in easy language to follow (maybe a glossary would help too).
Discussion of future funding options – Sarah Green (FR)
The meeting finished up with a discussion on further funding opportunities given that current project funding ends in March. There has been a no-cost extension until end of December 2019, but no further funds for staff time.
Sarah is putting together a proposal to the Defra future-proofing plant health (FPPH) programme 2019-2022. Within this she is looking to build a collaborative framework to support the continued development of accreditation. The feedback was that the timing was good for collaboration and the Phyto-threats project has a lot of information to feed into scheme.
Another part to the FPPH proposal is aimed at developing a standardised nursery testing protocol for metabarcoding to be incorporated into an accreditation scheme. This would use methods already developed as part of the project, but broaden the scope to include other types of pathogens such as bacteria. It was suggested that nematodes could be included too. It was agreed that community modelling for areas within the nursery prone to accumulation of pathogens would be useful.
On the question of the ability of metabarcoding to assist in nursery surveillance during statutory plant health inspections, the comment was made that, crucially, you need to isolate an organism at some stage. Also, results need to be fast, so staff can go back to the nursery almost immediately.
Another suggestion was that it might be possible to tie in nursery sampling with something else of service to the nursery, for example testing peat/water/ cuttings from Kenya! Even to ants or leaf-hoppers! Technology moves on too, the processing gets faster, better.
It was asked if Sarah had seen the HTA’s paper on how technology is being managed because this might give further guidance. Sarah confirmed she has had discussions with the HTA plant health lead and that he is supportive of the Phyto-threats project feeding into the Assurance Scheme.
Other thoughts on funding included developing something on the macroecology of Phytophthora and related groups. It might be possible to relate success in environmental/hosts/traits to host/pathogen relationships and latitudinal or elevational gradients i.e. what makes species successful? There could be added value of joining up research on trees with ornamental and agricultural research. The BBSRC standard grant round might be a good starting point, i.e. better predication on landscape-scale spread.
A comment was made in regard to the proposed accreditation scheme that the UK garden centre association has an annual meeting. They might be interested.
Two of the WP4 team have submitted a NERC DTP PhD proposal for continuing some Phytophthora genome work.
Another area for future consideration might be to identify where the unusual species have come from? What is the point of entry into environment? Traditional paperwork trail has been used as a tool. If you think about the nursery data, how does that Phytophthora come to be there? You can monitor sewage, fish farms, eDNA. NERC might provide support? It was also asked if there would be value in mapping Phytophthora species data for the UK? One of the bioinformaticians also works on a bacterium and is trying to identify through sequence analyses whether bacteria are transmitted vertically through seed or through the environment. They need to find the right bit of the genome, then they have to train the tool.
Following the end of this discussion the meeting was closed and Sarah thanked everyone for their contributions.
National Plant Show 2018
The Phyto-threats project and three team members returned to the National Plant Show again this year, held in Stoneleigh Park, Warwickshire, June 19-20th 2018. This is one of the largest plant trade shows in the UK, featuring over 160 exhibitors and receiving around 1400 visitors, representing garden centres and retail nurseries, as well as wholesale nurseries, online and mail order retailers, garden designers, consultants and local authorities.
The Phyto-threats stand had a poster display aimed at raising awareness of the link between the plant trade and Phytophthora outbreaks in the wider environment, and offered fliers with results from the consumer survey on ‘attitudes and behaviours of the UK’s plant buying public’ as well as some preliminary results from the ongoing nursery surveys for Phytophthora. The project also had a seminar slot on both days in which the nursery survey work was presented including Phytophthora findings to date and key messages so far in terms of management practices linked to high levels of Phytophthora infestation.
Once again the stand received a steady stream of interested visitors over both days which enabled some very useful networking opportunities. It does appear that awareness of pest and disease issues in trade is rising, largely due to concerns over Xylella, but also Phytophthora. Project team members took the opportunity to distribute a nursery and garden centre survey questionnaire aimed at improving our understanding of the supply chain; the perceived benefits (or not) of an assurance scheme; the basis of these attitudes (e.g. experiences of pests and diseases; use of measures for managing tree disease); and the willingness to pay extra or to travel further to buy accredited products. Information gained from this survey will be used to guide the development of effective accreditation. The survey is also available online.


Phyto-threats project team meeting April 2018
April 26th 2018 held at CEH, Wallingford, Oxfordshire
The aim of this meeting was to bring the entire project team and members of the Expert Advisory Panel together to share and discuss research progress since the last all-project team meeting on October 3rd 2017, and to outline and receive feedback on future research plans.
WP1 Phytophthora distribution, diversity and management in UK nursery systems – David Cooke (JHI), Leighton Prichard (JHI), Pete Thorpe (JHI) and Tim Pettitt (University of Worcester)
David Cooke introduced the entire WP1 team, including those involved in nursery sampling, processing in the lab, and analysing data, and he reminded everyone of the WP objectives and methods. He noted that processing of the filters from the water samples is very time consuming and that processing only the buffer solution that the filters are placed in might yield the same information with much less sample preparation time. They will be comparing buffer and filter samples from a subset of nursery samples to ensure that they are not missing any phytophthoras trapped on the filters.
For the fine scale sampling they have now obtained 49 sample sets from the 15 partner nurseries (6 in England, 1 in Wales and 8 in Scotland). Some results have now been reported back to all nurseries and this has promoted a more positive attitude among nursery managers towards the project. The WP1 team now have 2292 samples from 150 different host plant species, plus associated metadata. Of these samples, 1660 have been PCR tested for Phytophthora, including 617 from plant roots, 193 water filters and 850 buffer samples associated with water filters. They have also carried out isolation from some samples, yielding P. austrocedri, P. cambivora and P. lateralis. The team have been testing new methods for DNA extraction and have moved on to using kits rather than the phenol/chloroform method.
A question was asked about how sick plants were targeted – by eye?, or does the nursery manager ask the team to test specific plants? The answer was both, and that field notes and photos are taken of sampled plants so that results can be linked back to symptoms.
For the OPAL community sampling of local waterways, in cooperation with David Slawson and Vanessa Barber, the WP1 team have received 26 samples to date from three locations (north Wales, south Wales and Glasgow). These have been PCR tested for Phytophthora and about 50% are positive. This sparked some discussion on the usefulness of rolling out in the future the OPAL citizen science/public engagement element to a broader project looking at pathogens in the wider environment, particularly for early detection in sentinel plantings.
For the broad scale sampling, 64 nurseries have been sampled to date; 25 in Scotland and 39 in England/Wales. Samples usually involve 5-10 root samples per nursery. So far 408 root samples have been received. Root extractions have yet to be done on these samples. Sample packs for the 2018 broad scale sampling have been sent out to PHSI.
David then showed a range of photographs of plants sampled across nurseries and other general observations and reiterated that messages about management are being communicated to nursery managers. He presented a slide showing progress since the last project team meeting in October 2017, including data on the large number of samples processed during this time as lab testing has been accelerated. The first Illumina plate has been run and analysed, and results from the PCR testing and Phytophthora species data were reported back to nurseries in March 2018. David went on to show a number of graphs summarising PCR results by substrate and nursery and emphasised that data interpretation in relation to management practice is needed next.
There was a question on whether the sampler affects the number of positives, and that it might be good to see if certain people are getting higher numbers of PCR positive samples. The answer was that there is a fairly large team of people sampling most nurseries and that the team always includes at least one experienced plant pathologist. The sample team usually walks around the nursery first thing and decides together where possible which plants to focus on.
Another question was asked about whether the distribution of PCR positives and negatives could be plotted across nurseries to look at outliers, ie those nurseries with particularly high or low numbers of Phytophthora positives, and that could be related back to management practice. The discussion moved on to how the nursery sampling data can be analysed statistically, for example, how can we show that a particular practice results in a high Phytophthora load? It was suggested that the project establishes a small working group to decide how to analyse the data statistically.
David showed a slide summarising the Illumina run carried out with the first set of positive PCR samples. Samples containing synthetic control sequences at different concentrations were included on the plate to test the sequencing error rate, the indexing error to set an acceptable threshold read number (currently 50-100 reads) and the sensitivity range. David stressed that the species data obtained in this first sequencing run are preliminary and that full validation is required against a set of reference Illumina sequences obtained from 45 known Phytophthora cultures already set up on a control plate due to be run in May. This should provide important data on ITS1 sequence variation within species.
David then went on to show an example of the results files sent to the nursery managers back in March, including a cover letter explaining the methods and what the results are likely to mean. A brief comment was included on each Phytophthora species reported by the sequence data, ie its usual hosts and whether it is regarded as an aggressive species or not. David also discussed the Illumina findings and challenges – ie most species were Phytophthora, with some downy mildew or Nothophytophthora. There were low levels of Phytopythium in the sequence data. He ran through an overview of species found in some samples, for instance a high diversity of species particularly in river and puddle samples. Some species results were unexpected and we need a means of examining false positives. Reporting results back to managers is also challenging as the implications for management are not always clear. Summary findings will be reported back to the Plant Health Risk Group via Jane Barbrook of APHA. David also pointed out the dynamic nature of Phytophthora taxonomy, showing examples of the clade 6 phylogenies in 2003 (12 taxa) versus 2015 (30 taxa). He finished his presentation by outlining ongoing and future work; that the nursery sampling will be completed in 2018, many more Illumina plates need to be run and the need for data interpretation in relation to management practice, ie practices need to be related to Phytophthora findings.
A question was asked about relating DNA findings to actual viable propagules and that it would be useful to back up the DNA data by doing isolations/colony plate counts. This was addressed in part by a presentation from Tim Pettitt (below).
Tim Pettitt (University of Worcester) was able to demonstrate some results from his baiting and plating water samples conducted at a subset of nurseries sampled in this project. His data showed numbers of viable oomycete spores in different water samples that had also been PCR tested for Phytophthora. He recorded zero viability of oomycetes in a treated water sample (filtered and chemically treated) that was positive for Phytophthora in the PCR test. Therefore we do need to be careful in interpreting data because presence of DNA in a sample does not necessarily indicate viable propagules ie the PCR test is not a good indicator of viability. Tim also demonstrated his amendment to the water sampling methodology using a bike track pump used to push water through a bottle containing the water sample linked to several filters. This greatly speeds up the process of water sampling and reduces potential for cross contamination of samples. He uses bottles of fizzy water – they are sterile, and the fizzy water can be used to clean the filter heads and tubes, then replaced with the sample water.
Leighton Pritchard gave a presentation on the bioinformatics element of the WP1 work, entitled ‘Classification Performance Evaluation’. Through the use of sound tracks illustrating distortion of spoken phrases to represent the sequence ‘noise’ that the bioinformatics pipeline has to deal with when attempting to assign a sequence to a species, he demonstrated the importance of removing the ‘noise’ from a sample containing sequences (ie ‘noise’ meaning the sequences and sequence fragments that we are not interested in) so that the sequences are assigned to the correct species. He reiterated the importance of training and test sample sets, for example the 45 known Phytophthora species that will be Illumina sequenced so that we are clearer about which ITS1 sequences belong to which species. This will help us determine true positives and true negatives from false positive and negatives in our nursery samples.
The question arose about how much importance we attribute to distinguishing between closely related species. This would depend upon the species e.g. distinguishing P. rubi or P. fragariae would be not important. Sometimes it’s necessary to take a look at the sequences and determine manually. Some difficulties encountered include reference sequences that appear to differ, but are actually from the same isolate sequenced by different labs. Pete Thorpe discussed some of these conflicts in his presentation.
Pete Thorpe finished up the WP1 presentations by running through the metapy pipeline (to remind us of the methods and clustering tools). He also presented an update on the Phytophthora ITS1 reference database in which Sanger sequences have been obtained for 40 Phytophthora isolates and run through metapy to check the accuracy of the database. Some sequences did not cluster to the database, but did match 100% to sequences in GenBank; these entries were added into the database.
The performance of each clustering tool in metapy was tested on the Illumina sequence data generated from four samples containing mixes of DNA from known Phytophthora species. For each clustering tool Pete tested the sensitivity, precision, false negative rate and false discovery rate. Some species with very similar ITS1 sequences cannot be separated by most of the clustering tools e.g. P. capsici. Bowtie can separate them, but only if the ITS1 sequence is a perfect match with the database sequence. Basically Swarm performed best in terms of the above criteria, but none of the tools were perfect. Manual assessments are necessary when determining which species of a species cluster is most likely to be the one present.
Pete also talked about the error rates in the Illumina sequence output. Four random synthetic control sequences were synthesised with the same mean length and base composition as the Phytophthora database sequences, but with no BLAST hit and processed in the same way as the nursery samples. Errors can occur during PCR, through Taq error and during Illumina sequencing. He showed the frequency distribution of errors in the control sequences and where on the sequences the mismatches occurred. Errors also include indels and chimeras. The majority of the error variation occurred within two mismatches. Looking at the sequences, when the clustering threshold was set at 3 mismatches then P. ramorum was mis-identified as P. lateralis; at 2 mismatches, P. ramorum was correctly identified. Since a large amount of the dataset was represented within one mismatch of the control sequences a strict 99% threshold was used to cluster sequences.
WP2 Feasibility analyses and development of ‘best practice’ criteria – Mariella Marzano (FR)
Mariella started the WP2 presentation with an overview of milestones and outputs, and outlined progress so far, including the public consumer survey and resulting publication – a ‘glossy’ summarising the results of the public consumer survey which was produced for the THAPBI dissemination event in February. The online consumer ‘smart survey’ aimed at the public, nurseries, garden centres and landscapers has been distributed via horticultural magazines and email, although participation to date has been very limited. The WP2 team have been carrying out interviews with managers of the partner nurseries, liaising with FERA/HTA over the pilot assurance scheme, and liaising with FERA on the cost and responsibility sharing project, in which Gregory Valatin (FR) accompanied FERA economists during a number of nursery interviews to gain an understanding of the costs of different management practices.
The next steps for WP2 are to increase participation in the online ‘smart survey’. This could be done by using some of the budget to pay a company to conduct the survey for the project team. The WP2 team also need to improve economic data gathering through interviews with nursery managers. Interviews to understand purchasing habits and attitudes towards accreditation are also going to be carried out with retailers and garden centres, local authorities and other managers of large parks and gardens, as well as the landscaping sector. A series of focus groups will also be held in response to the interview and survey findings on appetite for accreditation.
Mariella ran through the questions asked in the nursery interviews and presented some of the findings so far in terms of nursery manager perspectives on disease threats, what they think about management for best practice, and what influences their plant purchasing decisions. She found that appetite for accreditation tended to be based on the size of the nursery and business objective. Some of the perceived benefits are that accreditation will provide reassurance to the customer as well as a training/‘safety net’ for the nursery (in terms of compensation). Participating nurseries would also be ‘seen to be doing something’ and it would allow traceability. Some of the perceived challenges are that there is currently little consumer awareness of the need for accreditation, the benefits of accreditation need to outweigh the cost of membership, and there was scepticism whether accreditation would change behaviours. Additionally, there is the common misunderstanding that we are trying to impose yet another scheme, instead of providing scientific evidence that would feed into a scheme. Not every nursery has got the message about disease threats, so engagement on this issue will be important, and how would accreditation be policed?
One comment was that there needs to be an update on progress of existing assurance schemes in the UK, looking to see how these could be run. Mariella confirmed that the WP2 team will resume engagement with the HTA and Defra over the pilot assurance scheme to ensure Phyto-threats project findings are used to influence the development of the scheme.
Mariella then held three discussion sessions. In the first session she asked the project team ‘what is ‘best practice’? She listed twelve nursery ‘best practices’ and asked if all of these should form the basis for accreditation and whether other best practices should also be included. This resulted in some discussion with one comment being there was a need for continuous monitoring of stock as part of accreditation criteria. This would require staff training. Another comment was that plant protection products can often ‘mask’ symptoms rather than solving the problem, and that this needed to be considered within accreditation.
In the second session project team members were asked to get into pairs to discuss which key questions the WP2 team should be asking of retailers, garden centres, local authorities and landscapers. Questions should include: What are their key suppliers? What is their biosecurity knowledge and experience? What are their purchasing practices, and what influences them? And what are their perceptions of consumer demand? One important point raised during this session was the need for biosecurity and plant health to be stipulated as part of the plant procurement process to allow purchasers to select what they consider to be the healthiest/the least risky bid rather than the cheapest bid. This needs to be taken on board by the government for example in setting plant procurement policies for local councils. During this session Mariella also asked for local authority, landscaper, garden centre and retailer contacts for interviews. Several members of the project team responded that they would be able to supply contacts, and even help with the smart survey in this way.
The third session dealt with focus groups. Three focus groups are required this financial year. These will involve small groups of selected stakeholders discussing questions around feasibility of best practice and attitudes towards accreditation. This could be done via the HTA and the pilot Plant Health Assurance Scheme; for example would they be influenced by the consumer survey? The WP2 team could also meet around APHA and retailers to have a session on best practice and accreditation. More discussion is needed on this.
Finally, Mariella presented some slides on behalf of Gregory Valatin (FR) who is looking at cost-benefits of best nursery practice. The objectives are to undertake an appraisal of the costs and benefits of options for developing best practice in UK nurseries to mitigate risks of further Phytophthora introduction and spread, both from the UK nurseries’ perspective and from the perspective of society as a whole. Mariella asked the team to consider the different scenarios that Gregory could use in his analysis, including the most appropriate baseline of best practice, which nursery characteristics to assume in the analysis and how to estimate cost of benefits to nurseries as a result of introducing best practice.
There was some discussion around whether an accreditation scheme should imply no use of imported seed. Current schemes vary on this issue, with the Woodland Trust’s UK Sourced and Grown Scheme not allowing importation of seed from outside the UK. Seed can certainly be a source of some pathogens. A baseline scenario should involve nurseries having implemented some, but not all, best practices, assuming some importation of stock from outside the UK. More time would be needed to explore these economics questions fully and it was decided that Gregory should speak directly to Tim Pettitt, Jon Knight (AHDB), and Jane Barbrook and Kelvin Hughes of APHA as experts in the nursery sector willing to offer advice.
WP3 Global Phytophthora risks to the UK – Beth Purse (CEH) and Mike Dunn (FR)
Beth Purse presented the WP3 work, starting off by reminding the project team of the WP objectives. For Objective 1 (Risk of Introduction) the aims are to identify the most important trade and recreational pathways linking Phytophthora source regions to the UK, to model introduction risk based on transport networks, source and destination characteristics and to test links between introduction risk and traits. One milestone was to compile a global country-level database of records of occurrence/arrivals of Phytophthora. This milestone is now nearing completion with 17,371 country level Phytophthora records, and 1417 species x country combination records obtained from various sources. These state where possible the source/recipient country, year of first record (linked to arrival?) and invasion status. There are of course large differences among countries in national recording and biosecurity effort and this is reflected in the number of records per country. In the analyses the WP3 team are looking at pre-2000 records as the potential ‘source’ distribution of Phytophthoras and post-2000 records as Phytophthora ‘arrivals’.
Preliminary analyses have been carried out using post-2000 Phytophthora records per country as the response factor and a set of predictors including trade connectivity to pest source countries (based on total imports of live plants), biosecurity effort (amount of invasive species legislation per country) and surveillance effort (number of official pest records from IPPC). Live plant imports and surveillance effort together explained 59% of the deviance in the number of new Phytophthora species recorded in a country since 2000.
UK Phytophthora records have also been compiled from a range of sources to enable models to be developed that relate Phytophthora species frequency of interception and extent of onward spread to biological traits. To assist with these analyses Beth held a short focus session in which she asked the project team to list what they felt were the most important factors influencing Phytophthora establishment in a new location, and rate of onward spread. These were then passed back to the WP3 team.
Beth then moved on to talk about the WP3.2 work (risk of establishment and spread) in which the team have continued to build the database of global Phytophthora records. They now have 11407 records for 82 species from 38 countries. In collating these data they have been prioritising countries that are climatically similar to the UK. Their niche models will predict potential global impact of Phytophthora species and the risk of their establishment and spread in the UK, excluding those species only known to occur in soil or water (ie no known plant host), species with no known woody hosts and species with non-relevant woody hosts (ie hosts not important in the UK). Their analyses will have to account for biases in reporting effort as developed countries have much higher species reporting than non-developed countries. This will be done by mining the scientific literature, oomycete and fungal databases (including GenBank) and adjusting weighting of records for species in highly recorded areas.
The Phytophthora traits’ database was merged in June 2017 with a similar database compiled by researchers in Australia and New Zealand. It contains data on 179 species and will likely be maintained in the longer term by Scion in New Zealand. A phylogeny will also be included in the database at some point. A publication is planned which will use the traits’ database as a conceptual framework for linking biological traits with invasion success of Phytophthora. Questions to be asked include whether closely related species share similar values for traits or groups of traits, or have traits linked to invasion evolved independently in several places in the phylogeny? They will also consider strength of phylogenetic signal in traits. Beth then ran through some traits which they have hypothesised to affect invasion success (ie survival structures, thermal tolerance, sporangial features). To do this they are using an ITS-based phylogeny (from Treena Burgess in Australia) for 179 species as well as two recent multi-gene phylogenies in order to resolve deeper nodes. Beth showed a series of slides with results from analyses so far illustrating strength of phylogenetic signal from sporangial features, reproductive traits and temperature traits, as well as rate of trait diversification over time – the latter showing high within-clade disparity suggesting rapid diversification and independent evolution to share common traits among clades.
Other questions being asked in the analyses are ‘do thermal traits especially cold tolerance modulate invasion of Phytophthora into temperate regions? (ie are emerging infections at higher latitudes linked to cold tolerance?) and how traits co-vary with each other (trait syndromes) and how much of this is driven by phylogeny? In terms of the global impact of Phytophthora the main question being asked is ‘can species traits explain the global impact of Phytophthora?. This analysis uses impact metrics including geographical extent (number of countries in which a species has been reported) and host range (number of known host families). These data are being compiled from various international databases. The team have also looked at whether trait syndromes outperform individual traits as predictors of global impact. Their findings so far suggest that root (and foliar) disease symptoms predict a broad host range, that trait syndromes are more ecologically informative about a species’ global impact than individual traits and that it might be possible to develop a traits based ‘early warning’ system for pathogens which have similar traits, but no impact yet.
Beth finished up by running through the plans for this coming year which are to submit papers on (i) the traits’ database and phylogenetic analyses and (ii) linking traits and global impact. They will fine-tune their species-specific trade models, develop UK and European spread models and finalise the niche models. They plan to finalise the model outputs with policymakers and practitioners in order to develop tools that people want, i.e.</em), interactive source maps and lists of Phytophthora spp. associated with key forestry species.
Mike Dunn presented on work done so far looking at tourism and recreation as a pest and disease pathway, using methods such as literature review, visitor data from susceptible tourist attractions and by conducting a survey of internationally based plant pathologists. The literature review has found numerous studies linking spread of a range of invasive organisms to tourism, including P. ramorum. A recent Visit Britain survey revealed 31 million people visited Britain over a 12-month period with a third of these visiting parks/gardens as a stated objective. Visitor data have also been collated from seven of the most popular parks and gardens in Britain, including weekly visitor data from Kew Gardens. The team are now going to look at tourism data to see if there are links between tourism and Phytophthora introductions in the UK. Sixty-one plant pathologists worldwide have responded to a survey which included among the questions whether they considered international tourism to be a potential pathway for invasive pests and diseases. Fifty-six of the respondents said yes to this question, which is backed up by data from several studies. When asked to rank levels of perceived threat of bringing in pests and disease, imported plants and trees was ranked much higher than incoming international tourists.
WP4 Predicting risk via analysis of Phytophthora genome evolution – Ewan Mollison (University of Edinburgh)
Ewan presented on the WP4 work done so far (since the WP started in August 2017) and began by outlining what can drive the evolution of a pathogen through intrinsic factors (ie duplication, rearrangement, insertion, deletion of DNA regions) and extrinsic factors such as hybridisation between species and transfer of genes between species. He ran through the aims of the work which are (i) to compare genes from available sequenced Phytophthora genomes in order to identify a core set of Phytophthora genes common to all species as well as species-specific genes, (ii) sequence the genomes of three less damaging species which are closely related to highly damaging species, so that genes involved in virulence might be identified and (iii) study target genes and gene families known to be important for virulence to identify how variations in these genes change pathogen behaviour such as host range and pathogenicity.
Ewan illustrated the sequencing and assembly strategy for P. austrocedri as an example of how a genome assembly can be improved. He found that 49% of the P. austrocedri genome consists of repetitive DNA. At this point Ewan was asked whether it would be possible to correlate species with high levels of repeat content with host range of the species – the answer was yes, it might be worth looking for associations with genome size and biological traits, although generally for other species genome size correlates with nothing!
Genome assemblies are now available for 26 Phytophthora species, all in varying stages of ‘finished-ness’. Most genomes are released along with predicted genes and protein sequences. Ten genomes have been released purely as scaffolded assemblies and gene prediction will need to be carried out on these. Ewan presented a graph showing large variation in the number of predicted proteins (over 30 amino acids in length) among currently available genomes, with the caveat that the protein data are likely to be an over-prediction and that the gene-prediction tools will need to be refined. A graph showing assembled genome size versus repeat content also illustrated the high levels of repeat content in certain genomes such as P. alni (hybrid), P. cambivora (putative hybrid) and P. infestans. Larger genomes are often a result of expansion of repeat regions, with these repeat regions often evolving rapidly which is very useful for overcoming host resistance.
Ewan assessed the completeness of coverage of each of the 26 Phytophthora genomes by looking for the presence of 234 ubiquitous genes expected to be present in all species. His findings showed that many genes may be missing from an assembly. For example 22/26 genomes were estimated to be 90% ‘complete’, 3/26 over 70% complete and the P. alni genome only 37% complete. Ewan then looked for orthologous genes present in all 26 genomes and found 2,107 genes or gene clusters common to all genomes. A phylogenetic tree inferred from these ‘core’ genes split some of the species from the same clade (ie species in clades 1, 3 and 8) however much more work is needed to refine this analysis.
Ewan then described some analyses done using an example gene family; the xylanases (xyn), which are cell wall degrading enzymes which specifically target hemicellulose, an important constituent of plant cell walls. There are four major xylanases which have been identified in Phytophthora; xyn1, xyn2, xyn3 and xyn4. Sequences from all xyn genes found in the 26 Phytophthora genomes were aligned, and phylogenetic trees constructed for each gene. The xyn1 and xyn2 genes grouped into two distinct clades whereas the clades were less clearly defined for xyn3 and xyn4. Looking at the presence/absence of each gene among the 26 Phytophthora genomes revealed that not all species contain all four xyn genes which may be due in part to incomplete assemblies, and also that xyn sequence differences occur in some species which results in their falling outside the expected clade groupings for that particular gene.
Ewan finished his presentation with a cautionary tale; strangely anomalous xyn gene sequences were downloaded for P. taxon totara because the source database had been mistakenly linked to a P. kernoviae genome download. Always check the source!
Further WP4 work will involve sequencing P. obscura, P. foliorum and P. europaea to add into the analyses, together with any newly available Phytophthora genome assemblies. The xylanase gene family analysis will be expanded and other gene families of interest will be investigated ie RXLR effector proteins.
WP5 Synthesis and integration – Sarah Green (FR)
Sarah Green rounded off the meeting with a short overview of WP5 activities since October 2017, including Board meetings and the reports/research summaries recently posted on the project website as well as the successful uploads of all project outputs and outcomes to ‘Researchfish’. There was some discussion on stakeholder engagement activities planned for the coming year. It was decided not to attend the National Plant Show this year, but rather to wait until next year to present project results. There was agreement for the need to engage more formally with Defra and the HTA over the pilot assurance scheme to ensure that the Phyto-threats project data can help to shape this scheme. The next stakeholder workshop will have the theme of ‘securing resilient outcomes – scoping the potential of an accreditation scheme and building a framework for its continued development’. It was thought that delaying the workshop until spring next year in order to have a more complete set of results to present would not fit in well with the nursery timetable – spring is very busy. So a decision will be made soon on whether to stick to the same format of holding the project team meeting followed by stakeholder workshop over two days in October. This will be decided at the next Board meeting.
IUFRO meeting
Phyto-threats attendance at the 8th Meeting of the International Union of Forest Research Organisations Working Party (IUFRO) 7.02.09, Phytophthora in forests and natural ecosystems.
Sapa, Vietnam 18-25th March 2017
Sarah Green (Forest Research) attended the 8th meeting of IUFRO working party 7.02.09, ‘Phytophthora in forests and natural ecosystems’ held in SaPa, Vietnam at the end of March 2017. This meeting, which was attended by ~100 forest Phytophthora researchers from around the world, began with a two-day pre-conference field tour which was spent visiting forest nurseries and plantations of Eucalyptus and Acacia affected by Phytophthora cinnamomi. The nursery operations viewed during the tour were less than optimal in terms of Phytophthora management, but moves are afoot in the country to educate nursery managers and improve disease management practices.
During a conference session on Dispersal Pathways/Urban Horticulture, Sarah presented an overview of the Phyto-threats project, including a series of slides prepared by Louise Barwell and Beth Purse (CEH) on the global Phytophthora risk modelling approaches they intend to take, finishing up with a request for researchers to contribute their Phytophthora records for the development of global niche models. Several Phytophthora researchers from a range of countries have already contributed data, or are planning to do so, and resultantly the Phyto-threats modelling team are having good success in pulling these distribution data together (more than 7000 records to date).
In a closely parallel project, Peter Scott of Scion, New Zealand, demonstrated his modelling approaches to predict global Phytophthora diversity and biosecurity risk. His time series models predict up to four times more Phytophthora species than those currently described. Peter and his group are also modelling Phytophthora invasiveness traits to predict global risk. Given the degree of overlap in the two projects, and thanks to a surprisingly good Wi-Fi connection, a successful Skype meeting was held one evening between the New Zealand/Australian Phytophthora modelling team (plus Sarah) located in a hotel bedroom in northern Vietnam and Phyto-threats modellers’ Beth Purse and Louise Barwell located in an Oxford office!. Future meetings are now planned between the two groups to scope out potential areas for collaboration. This may include sharing of Phytophthora traits’ databases independently developed by both groups with a proposal to produce a combined traits’ database for every known Phytophthora species that will be published online and managed and updated through a collaborative effort.
Another major conference topic of interest to the Phyto-threats project was the use of metabarcoding to detect Phytophthora species in soil and water in a range of different ecosystems around the world, including plant nurseries. Some Phytophthora species appear to be ubiquitous across highly differing environments, and as the global picture continues to develop it may be possible to see (and hopefully understand) patterns in species distribution and diversity. Miguel Redondo based in Uppsala, Sweden, reported on a study of Phytophthora communities in eight nurseries, 16 rivers and 14 forests in Sweden, and undertook modelling to identify functional traits associated with establishment. Their approach was particularly interesting for having included both highly managed and less disturbed sites in a single study, and the authors observed large differences in the composition of Phytophthora communities in the three environments. David Cooke (JHI), who leads Phyto-threats work package 1, gave a presentation on methods used for sampling Phytophthora diversity which highlighted some of the pitfalls of using the multi-copy ITS region for Phytophthora identification. David described a bioinformatics pipeline developed by project colleagues’ Leighton Pritchard and Peter Thorpe (JHI) which uses different clustering algorithms to address within-species sequence variability in species assignations. This pipeline, a key output from the Phyto-threats project, will soon be made publicly available for metabarcoding analyses.
In terms of Phytophthora management in nurseries, a critical link in the dissemination of Phytophthoras globally, Laura Sims of the University of California presented a nicely contrasting ‘before and after’ study showing how a series of changes to nursery management practice can dramatically reduce Phytophthora detections. Similarly, Agnes Simamora of the University of Nusa Cendana in Indonesia showed how P. boodjera infections in a containerised production nursery can be eliminated by steam-sterilizing the re-used seedling trays. Such evidence of the effect of relatively straightforward management changes on reduction in disease load will be useful in upcoming engagement activities with UK nursery managers planned as part of the Phyto-threats project this year.
National Plant Show 2017
Stoneleigh Park, Warwickshire, June 20-21st 2017
Phyto-threats project had a stand and seminar slot at the National Plant Show, held in Stoneleigh Park, Warwickshire, June 20-21st 2017. This is one of the largest plant trade shows in the UK, featuring over 160 exhibitors and receiving around 1400 visitors, representing garden centres and retail nurseries, as well as wholesale nurseries, online and mail order retailers, landscapers, garden designers, consultants and local authorities. The purpose of the Phyto-threats stand and seminar was to raise awareness of the link between the plant trade and Phytophthora outbreaks in the wider environment, and to inform growers of the role they can play in helping to reduce the spread of Phytophthora through best management practice. The stand received a steady number of interested visitors on both days, with a good number of these visitors willing to participate in an online survey designed by members of the project team to assess perspectives on accreditation based on best practice. It was also very useful for the project team to gain a greater understanding of what drives the plant trade, and how trade networks work across Europe and beyond.
Phyto-threats project team meeting May 4th 2017
James Hutton Institute (JHI), Invergowrie, Dundee
May 4th 2017
The aim of this meeting was to bring the entire project team and members of the Expert Advisory Panel together to share and discuss research progress over the first year of the project for the three work packages that have started, and to outline and receive feedback on future research plans. Following the talks and discussion sessions participants were given a short tour of the JHI pathology and sequencing labs.
The meeting started with a welcome by Sarah Green (Forest Research, FR) and brief introductions of everyone present including their affiliations.
09.30-11.00: WP1 Phytophthora distribution, diversity and management in UK nursery systems – David Cooke (JHI) and Pete Thorpe (JHI)
David Cooke introduced the WP1 team, which consists of David Cooke, Leighton Pritchard Peter Thorpe, Eva Randall, Beatrix Clark (all JHI), Sarah Green (FR), Debbie Frederickson-Matika (FR), Tim Pettitt (Uni of Worcs), Alexander Schlenzig (SASA) and Jane Barbrook (APHA).
Nursery sampling
For context, David provided a snapshot of the distribution of P. infestans clones in Europe from 2013-2016 based on 5000 isolates from the EuroBlight project, showing how an airborne pathogen can rapidly change its distribution. David then gave an overview of WP1 objectives and methods for the nursery sampling, including some practical issues such as range of plant species sampled, which varies among nurseries according to what stock they hold. Generally, the sampled plants are a mix of known and unknown hosts for Phytophthora, and may be symptomatic or asymptomatic at the time of sampling. Mostly root samples are taken, except in a few instances where foliage/stem samples have been taken from symptomatic plants. Water supply is sampled at source and run-off. In general, sampling effort at each nursery is a balance of time available and the need for detail, focusing on working through the hazard points – ie incoming plants/water/pots/ground.
So far the team have carried out 18 sampling missions at 15 nurseries (6 in England, 1 in Wales, 8 in Scotland). This has resulted in 1009 samples (everything in triplicate) including plant roots (93) from a range of 35 hosts, 132 water filter samples and 170 samples of buffer associated with each filter. Water samples came from water washed through plants, boreholes, ponds/ditches, equipment washing (e.g., trolleys) and water control blanks. To date, 395 samples have been PCR tested for Phytophthora. David described some of the necessary changes to lab protocols to improve the efficiency and accuracy of processing.
Data were presented showing the number of Phytophthora-positive samples for 10 nurseries (labelled 1-10 and not named in the presentation). Sample types were broken down into water filter, water buffer and roots. There were clear differences among nurseries in relation to the number of Phytophthora-positive and -negative samples, and this could be related to practices observed during the sampling missions. David did outline the issues of working with each sample in triplicate, e.g., when 2 samples are negative for Phytophthora and 1 is positive, and how they deal with such results.
There was discussion on the different nursery sizes and practices, and how that relates to the number of findings per nursery as this should be taken into account when presenting data. Hard evidence is required linking practice to Phytophthora findings. Mariella Marzano (FR) made the point that a survey of management practices accompanies each nursery sampling visit so that a basis of best practice/worse practice can be developed in relation to Phytophthora findings. Participating nurseries are willing to know their results in order to improve their practice. Nursery data include turnover/plant species sold to help interpret findings. Detailed (fine-scale) sampling is only done for nurseries that volunteer. The question was asked whether sampling was designed to avoid bias, i.e., if you focus on a sick area of plants within a very large nursery you will bias the data. This is more a problem for plant samples than water samples. David Cooke confirmed that detailed notes accompany each sample collected, including whether samples were collected on a random basis or due to presence of disease symptoms.
A broad-scale survey is due to start soon in which plant inspectors will be collecting root samples for the project from nurseries during their routine Plant Health inspections. In particular it will be important to clarify with inspectors where and what they should be sampling. This element needed further discussion between APHA and JHI. David made the point that the aim of the broad scale survey is not to repeat the statutory testing, but rather to look more widely across nurseries for a different insight that will help inform the goals of the Phyto-threats project.
It was suggested that nursery samples are collected from areas not expected to have disease (for example where plants are apparently healthy), just to check what Phytophthoras might be ‘hiding’. In such cases the foliage could look healthy, but perhaps the pathogens are present in the roots or soil. Some Phytophthoras such as P. gonapodyides could just be ‘root nibblers’ and not viewed as pathogenic. The consensus was that it is still useful to know the distribution of non-aggressive Phytophthora species to see how they are distributed. The presence of these species might indicate a route in for other more pathogenic Phytophthoras.
It was asked whether information is available on where nurseries with Phytophthora-positive samples have sourced their plants, ie from plant passport numbers, and whether such information could be used to know what pathogens are in those areas where plants are imported/bought in from ?. David Cooke responded that data on plant sources are with each nursery and could be obtained from some of them, however, the true origin of the plants might not be known by their plant passport number. The length of time that plants have been in a nursery is a factor included on the nursery questionnaires.
David Cooke listed the Phytophthora findings to date by sample type, showing a higher proportion of positive samples from roots than water, although this would reflect the fact that sickly plants are targeted for the root sampling. Different production methods included;
- Water sources; mostly borehole, some mains plus rainwater and river water (‘filtered’).
- Plants in cells/pots above ground or on the ground, or bare root grown in the ground (ie forestry nursery).
- Production systems included wholesalers / holding / importing to production on-site.
- Premises included garden centres / nurseries / mixed / horticulture / hedgerow / forestry tree growers.
- Generally good plant health awareness was observed, but fungicide use was widespread.
David then showed examples to illustrate sampling points, such as water sources and types of plants sampled.
In terms of the next steps for the nursery sampling; David Cooke is to discuss the broad scale sampling with Jane Barbrook (APHA) and Alexandra Schlenzig (SASA), as well as co-ordinating the OPAL sampling with David Slawson. For OPAL, water samples from streams and other waterways will be collected by OPAL community volunteers in Plymouth, Cardiff, North Wales and Glasgow. David Slawson emphasised the need for good photographs and descriptions of sampling methods for the volunteers, and also expressed the need for rapid feedback on OPAL results to enthuse and engage OPAL volunteer interest. The fine scale sampling of the 15 partner nurseries will also be repeated in June/July and again at the end of the year. A question was asked whether root samples are also taken from plants sampled by water flow-through (ie plants are placed on a tray and watered to capacity, left to stand for 30 mins or so, and the water flow-through in the trays then sampled). If so, this would provide an interesting comparison of the two methods in terms of Phytophthora detection. David Cooke said that where possible roots were sampled from plants also subject to flow-through sampling, although sometimes time constraints prevented this. At the moment there is also a backlog of samples requiring DNA extraction from the previous round of sampling and getting this done will be a priority.
The first 176 Phytophthora-positive samples will be run through Illumina sequencing in May 2017. This will produce 15M barcode reads=156K reads per sample; each read approximately 250bp in length. Synthetic control samples will also be included in each sequencing run as a test of error rate.
Bioinformatics analyses
Pete Thorpe (JHI) presented on the bioinformatics pipeline that he and Leighton Pritchard (JHI) have developed called METAPY – this pipeline is a key output from the project and will soon be publicly available on GitHub for use by the wider scientific community. Pete began by describing the system for Phytophthora identification from sample collection, to DNA extraction, nested PCR of the ITS1 region with Phytophthora- specific primers, DNA library preparation, Illumina sequencing and analysis of sequencing reads by the bioinformatics pipeline, METAPY. Pete explained that other pipelines tend to use one clustering tool (ie to align sequence reads to a reference sequence in a database), however, each clustering method can vary and so METAPY produces individual results for five different clustering methods (BLASTClust, Swarm, V-search, CD-hit and Bowtie) so that results can be validated by comparing the different methods.
Pete ran through the different clustering methods and showed results from a control sample containing a DNA mix of ten known Phytophthora species. He explained reasons for variation and false-positive results for each clustering method. BLASTClust was the least discriminating method and reported 27 species; many of these false-positives were species in the same clade. Bowtie, on the other hand, only detects species with a 100% match to the reference sequence in the database and reported only 7 species in the test. CD-hit found 20 species, Swarm 16 and Vsearch 16. The reference database is yet to be adjusted for species which have highly similar or matching ITS sequences and this might sort out some of the false-positives. METAPY can also be set to error-correct reads before identification (Illumina can be prone to error – these sources of error are known and can be automatically corrected). METAPY is currently used for the ITS1 region, but can be edited for other genes.
Pete then described the main issue with using the ITS region which can exist in 50-500 copies in the Phytophthora genome based on assessment of ITS copy number using qPCR. The short reads produced by the sequencing method can’t resolve repeat/repetitive regions and so these get collapsed to consensus sequences. Perhaps a different sequencing method such as PacBio, which produces longer reads, will help to resolve this. Pete is re-assembling ribosomal DNA regions for all Phytophthora genomes to determine copy number and within-genome variation.
The question was asked how Phytophthora hybrids might be identified using the bioinformatics method. David Cooke agreed that chimera detection in the pipeline might throw them out and thus identifying hybrids will be tricky. Ana Pérez-Sierra (FR) mentioned the POnTE project which is assessing both ITS1 and the Cox region for Phytophthora detection to see how resolution compares. For other fungi, for example powdery mildew, the single copy B-tubulin gene is used for barcoding.
It was also asked whether the different ITS sequences present within an individual would show up in the metabarcoding data given the very high number of ITS reads produced with Illumina, so that within-species variation would be identified. Or, is it only the predominant and more abundant ITS sequence that gets resolved?. Sarah Green (FR) described the scenario with P. austrocedri that had arisen in a previous metabarcoding study in which some samples yielded high numbers of reads of an ITS sequence typical of the UK lineage plus a few reads of an ITS sequence typical of the Argentinian lineage. Initially it was thought that the Argentinian lineage might be present in Scotland – however, subsequent analysis of the P. austrocedri genome revealed that the UK lineage also has the Argentinian-type ITS sequence in the repeats, probably at lower copy number. David Cooke responded that some species do have multiple ITS types and he wants to build them into the reference database.
Beth Purse (CEH) commented that the WP3 team are receiving data on global Phytophthora distribution based, in many cases, only on ITS sequence detection. Ca we be confident in these data?. David Cooke replied that whereas some species have identical ITS sequences and cannot be resolved, the majority of Phytophthoras can be distinguished based on ITS. Taxonomic irregularities can also cause difficulties. The WP3 team will need to have clarification over which species can be confused so that they can mine the data that they have compiled. Can they trust the Blast analyses used to identify species?, and is there a timeline for species identification?, i.e., before a certain date the identifications would have been based on morphology alone and thereafter more likely to have resulted from mainly molecular tools or a combination of both. David Cooke replied that from about 1998 onwards, Phytophthora researchers were using molecular tools, and thus data are more likely to be reliably discriminated. The trick is to avoid over-interpretation. We also need to take care not to interpret previously uncharacterised sequences as new species just because they are not on the reference database. There was then a discussion/comment that ideally we should know the range of ITS ambiguity in each Phytophthora genome so it can be incorporated in the analyses. The existing reference database of Phytophthora ITS genomes is still being worked on and accuracy of the database is key to the outcome of analyses. Currently it is believed by the team that METAPY gives the best likelihood of a given Phytophthora species being present compared with other pipelines.
11.00-12.30: WP2 Feasibility analyses and development of ‘best practice’ criteria – Mariella Marzano (FR) and Mike Dunn (FR)
Mariella began with an overview of the social and economic research being carried out by herself, Mike Dunn, Gregory Valatin (all FR), Colin Price (contractor economist) and Tim Pettitt (Uni of Worcs, nursery engagement) and she reminded the project team of the WP2 objectives, listing milestones and outputs. Essentially this WP has three key parts; i) social analysis to assess applicability of nursery best practice criteria, ii) cost-benefit analysis of implementing best practice and iii) which elements of best practice that should underpin an accreditation scheme.
So far the WP2 team has mapped the stakeholder networks and created a stakeholder database. The consumer survey was supposed to be done in year 2 of the project, but since there was early interest in this work the survey was conducted in year 1 (more detail to follow). The team have done some context building; interviewing science team members and members of the Expert Advisory Panel to get a sense of what is needed, and Mike Dunn (FR) has also joined the WP1 team on nursery sampling visits. They are exploring existing values within the sector, experiences, and practices on disease and management. They are planning to conduct interviews and undertake participant observation at nurseries starting this summer. One of the key factors to assess will be potential attitudes and willingness to join an accreditation scheme. The team will also do wider industry focus groups, but they have yet to finalise the methods. They will be led by the data as it comes in. More surveys are being developed for landscapers, nurseries and garden centres, including supermarkets and superstores. They will prepare questions on the supply chain, disease threat perspectives, current management, policy tools etc and obtain information on decision management, where can and can’t nurseries change, perspectives for future management and willingness for an accreditation scheme.
Consumer survey
Mike Dunn (FR) gave a descriptive overview of results obtained very recently from the consumer survey which involved 1500 people. The data have yet to be analysed statistically. Mike ran through the questions addressed by the survey and the demographic data for the respondents who were typically members of the public who buy plants for their own home. The 19 survey questions were compiled and sent for approval by the project’s ethics committee before the survey was sent out. The survey was undertaken by a survey company (Toluna) and it targeted buying habits, perceptions of the seller’s biosecurity, and attitudes towards accreditation. The 1500 respondents were generally representative of the nation’s demographics, although 57% were aged 55 or over.
Survey results
Most people buy plants 1-2 times per year, most frequently sourcing plants from garden centres/DIY stores/supermarkets. Mariella noted that the project has not engaged these sectors yet and doing so will be important. Plant quality, cost and range were the top three factors in a consumer’s decision about where to buy plants from. Evidence of good biosecurity practice by the business came out as a fairly low priority. When it comes to selecting plants to buy, plant appearance, suitability for the intended site and cost were most important. Provenance and origin of stock were very low priority for consumers.
In terms of general awareness of pest and disease threats to plants; 10% of respondents had never heard of the problem, 62% had heard of the problem, but didn’t know much about it, 24% had heard of the problem and felt reasonably well informed and 3% felt very well informed. Respondents were assessed for their perception of the level of risk of different potential pathways for pest and disease introduction; online and mail orders were perceived as higher risk whereas consumer’s self-grown plants, specialist plant suppliers and nurseries gave most confidence as being of low risk. In terms of willingness to support an accreditation scheme; 9% of respondents chose not to buy accredited products because of the increased cost, 32% gave accredited sources little thought when making a purchase and 38% bought accredited products in some cases because they believed in the goals of the scheme. In terms of a potential accreditation scheme for plant sellers, 25% of respondents agreed with the principles of such a scheme, but had concerns about the cost being passed on to consumers.
The consumers surveyed spent on average £100 per year on plants, and 45% were happy to travel further (up to 160 miles, with an average increased willingness-to-travel distance of 26 miles) and 39% were willing to pay more (on average 18% more) for accredited plants.
Overall, the results of the consumer survey indicated a general willingness to support accreditation (within reason), but it also highlighted a need for consumer engagement about the risks of spreading pests and diseases.
Discussion of consumer survey
Mike’s presentation generated a number of questions such as ‘are attitudes to accreditation associated with i) a greater knowledge of pests, ii) experience with existing accreditation, iii) number of plants purchased, iv) types of plant sources, v) demographic factors ?.’ For example, it was asked whether the previous experiences of the consumer affected their attitude towards accreditation, ie if trees had been felled in their local area then is this likely to have increased their awareness and concern of pests and diseases?. The response was that, based on previous surveys, people generally don’t notice damage to trees in their area. The point of this survey was to find out where people buy their plants and what influences their behaviour. The OPAL project, involving community volunteers including schoolchildren, might help with raising awareness of younger generations. Another question was raised whether the behaviour of online respondents who completed the survey could be expected to differ from the public at large. The answer from Mike Dunn was that, overall, he is happy with the survey sample as it targeted a representative population in relation to location across the UK, gender and age bracket, bearing in mind that older people tend to be more likely to buy plants than younger people.
Next steps
A survey will be designed for the forestry industry and landscapers, and the project team and Expert Advisory Panel members will be asked for their inputs in shaping the survey questions. For the nursery and garden centre survey it is recognised that these groups can be both sellers and consumers. Mariella will also link in with the Horticultural Trades Association (HTA) who are working in partnership with nurseries to develop an assurance scheme.
The comment was made that growers need clear plant health advice on which to base best practice and that any assurance scheme should include management practice guidelines. The question was asked whether the HTA-piloted scheme was just for UK growers or international. Mariella responded that the scheme was for the UK market, but the HTA would like it to extend internationally. The scheme is being piloted now and could possibly be launched next year.
Economic analyses
Mariella presented a number of slides on behalf of Gregory Valatin (FR economist researcher) who could not attend this meeting. Gregory will undertake an economic analysis of consumers’ willingness to pay and travel for accredited stock. Gregory will look at cost-benefits of other schemes, on potential impacts of new Phytophthoras entering the UK and potential costs of not having best practice thus increasing chances of new diseases establishing. Gregory had a number of basic questions for the project team and advisory panel including;
- What kind of accreditation scheme shall we focus on?
- What is the source of additional costs-holding plants/use of fungicides or pesticides?
- Timescale for introduction of scheme?
- To what extent are nurseries expected to take up scheme?
- Timescale for uptake?
The team will consider these questions and report back to Gregory.
Sarah Green (FR) commented on a study from Californian native plant nurseries supplying restoration plantings showing how Phytophthora infections were greatly reduced after the nurseries implemented a number of key management changes. Although there were costs to implementing these changes, the resulting benefits were clear, ie less Phytophthora and less risk of passing disease to native landscapes, so reducing costs associated with outbreak eradication.
The question was asked ‘if we don’t have a scheme, what would happen?’. The answers were that there must be data on the costs of remediation following outbreaks. Obtaining such data could be tricky with some outbreaks being public and some private. Essentially, we don’t really know the baseline costs of disease eradication efforts. It may be possible to look at past trends and establish how action could level off the need for intervention.
The discussion returned to the topic of uptake of accreditation by nurseries; if a scheme involves too much paperwork it may restrict uptake. How do we bring growers on board?. Smaller nurseries will find it costly and not participate.
Gregory’s work will assess the anticipated effects of an accreditation scheme on introduction of Phytophthoras, their spread in the UK, effects on introduction of other pests and diseases as well as spread of pests and diseases already present in the country.
A last question posed: ‘What are the differences likely to be, compared to baseline risks, and how do we quantify them ?’ e.g., % reduction in time of species x in year y.
Glyn Jones (FERA) presentation on Future Proofing for Plant Health (FPPH) project
Glyn presented on a couple of economics-related projects he is working on. One is funded through the DEFRA FPPH programme and is looking at cost and responsibility sharing, modelling nursery networks and assessing markets for plant health and biosecurity. He is considering the public good characteristics of biosecurity, ie market failure if nurseries won’t provide the biosecurity that the public wants. Glyn presented a figure showing nursery sector complexity and the impact of poor biosecurity, and the difference between a pest staying put or moving into wider environment.
Glyn commented that we are living in changing times and ran through traditional government practices and instruments for intervention, including inspection, monitoring and surveillance, quarantine and pest exclusion. Research funding allocations reflect budgetary pressure from increased and changing risks. He asked if the industry is making a move now due to concerns over Xylella and the impact of Chalara. How should an accreditation scheme be designed? The situation is currently dynamic, with various Woodland Trust, Grown in Britain and HTA assurance schemes. The Woodland Trust want clean sourced trees, but there is a potential problem with the source, for example if the order is large then trees might have to be sourced elsewhere (ie outside the UK). Glyn also ran through a number of possible economic mechanisms to support an assurance scheme, such as i) cost sharing for outbreak control measures, ii) insurance (easier if the pathogen is defined and involves a small defined group of participants), iii) environmental bond (ie growers pay money, and if no disease they get reimbursed – this may have a positive impact on behaviour, iv) retrospective levy, v) cost sharing for risk reduction measures.
Interviews have been undertaken by Glyn and his team with some of the key players in the industry. There were some criticisms of accreditation/assurance schemes, in that if a scheme were voluntary you would likely only get growers joining who already operate best practice. If a grower were to be part of a scheme would they be subject to fewer Statutory inspections?. Generally, the idea of reduced inspections was not supported by industry, they actually like being inspected!. Questions that arose included why would growers join a scheme ?, what are the benefits to them?, what do you do about those growers who don’t join?. The feedback from growers was that any scheme would need to be government-endorsed and clear, with a strategic direction. There needs to be leadership from DEFRA and interaction with policy. The responses to malpractice need to ‘have teeth’ to incentivise adoption of good practice.
In terms of UK biosecurity policy, proactive temporary import bans of high risk plant material are hard to achieve. It was suggested that there should be expansion of nursery licences and inspections to other importers, for example landscapers and landscape architects. One possibility would be to base border inspections on the biosecurity rating of exporting nurseries.
In general, growers supported the notion of fewer rather than more assurance schemes and the idea of earned recognition, in that there would be benefits to members of such a scheme. There can be market distortion and there is often a mis-match between demand and time to supply. Growers said that there needs to be increased distinction of UK-grown material and a transition to more home-grown, and perhaps more co-operative working of growers in order to achieve required supply. There needs to be confidence in the domestic market.
In terms of future FPPH work, Glyn said that he would be integrating with the LWEC Phyto-threats project and would liaise with Mariella and Gregory. He will be conducting more interviews of those involved in trade and undertake a demand-pull analysis, modelling design and required uptake of an assurance scheme. Glyn also mentioned the RAPID trade project, funded by BBSRC and US National Science Foundation. As part of this project they have been modelling high biosecurity trade networks following pest/disease outbreak scenarios.
It was asked whether a UK-wide accreditation scheme should be compulsory. There followed some general discussion of this along with suggestions of a licensing scheme (which might just benefit the largest operators) and whether public benefit could be used to influence what public funds could go into the scheme.
Phyto-threats project team meeting May 4th 2017 (PM session)
13.00-14.30: WP3 Global Phytophthora risks to the UK – Beth Purse, Dan Chapman and Louise Barwell (CEH)
Beth began the WP3 presentation by reviewing the work package objectives; (i) risk of Phytophthora introduction, ii) risk of Phytophthora establishment and spread and iii) horizon-scanning for emerging pathogens (including assessment of tourism pathways of spread). The work package team comprises Beth, Dan Chapman and Louise Barwell (CEH), Ana Pérez-Sierra, Mariella Marzano and Mike Dunn (FR).
WP3.1 Risk of introduction
Beth, Dan and Louise have been compiling a global database of Phytophthora country-level occurrence for use in predicting risk of arrival in the UK for those species not yet present in this country. Dan and Louise will be looking at trade routes linking Phytophthora source countries to the UK. The global Phytophthora occurrence database so far includes 5838 country-level records and 1208 Phytophthora species x country combinations. The data include year of arrival (or rather 1st record in a particular country) as well as invasion status and state of establishment and spread. Data have been sourced from 5 databases: daisie/ ncbi/ gbif.org/ cabi/ westerdjik (formerly the CBS culture collection).
Dan Chapman presented on trade networks. In terms of trade pathways the focal commodities are horticultural/agricultural/forestry products. Dan has already completed an analysis framework for correlating trade networks to plant pest invasion patterns in Europe, and has looked at the best-fitting model for invasion (paper currently in press in Global Ecology and Biogeography). He has analysed national-level introduction patterns of over 400 non-native species and found that these were best explained by a model incorporating pathogen occurrence in source regions, climatic similarity between source and sink regions, and connectivity of these regions through multiple trade networks. For more recent invasions, the live plant and forest products networks gave the best model. Dan and Louise will now take this further with Phytophthora species, by adding new trade data and more refined pathogen traits that might influence invasion risk. Louise asked for thoughts about how Phytophthora traits might affect species’ abilities to exploit specific commodity pathways such as live horticultural plants, cut flowers, fruit, vegetables, soil, agricultural seeds, roots, food and feed, forestry products such as fuel, wood, and travel/ tourism, and to exploit new geographic regions with similar environmental conditions.
There followed some discussion on how Phytophthora traits relate to risk of importation and what the team are hoping to find. Beth responded that they would like to correlate certain traits or groups of Phytophthora with these traits as risky. They would then see where these ‘high-risk’ Phytophthoras occur and identify routes correlating with volumes of trade. Richard McIntosh (DEFRA) commented that there is a policy demand for this sort of information, including information on what countries are deemed to be of lower risk so can trade can be directed towards those countries.
Beth asked for sources of data on trade interceptions and traits of the organisms intercepted. Europhyt (European Union Notification System for Plant Health interceptions) was mentioned, although the team had already looked at the Europhyt database and didn’t find any Phytophthora hits. APHA/FERA have an extensive database of nursery interceptions in England and Wales, but mining it might be a problem as the database is so large. The WP3 Beth team can have access to the database as long as they know exactly what they want and the time period. SASA have already passed on the nursery interception data from Scotland to Beth’s team.
WP3.2 Risk of establishment and spread
Beth spoke about the potential environmental drivers of Phytophthora distributions, matching patterns of occurrence with environmental data. Niche modelling will be done on site-level occurrences (rather than country-level). They have amassed a global dataset of 8067 site-level Phytophthora occurrences, covering 81 Phytophthora species in 38 countries from a variety of data sources including culture collections, unpublished data from researchers in the Phytophthora community, governmental organisations and the literature. Due to recording effort the global distribution of these records is patchy and high-impact species are more frequently recorded. For example the P. cinnamomi dataset contains 3655 location records in over 20 countries. The agricultural sector is poorly represented so far, but Beth and Louise would like to ensure that the dataset captures the distribution in all relevant sectors to get a complete picture of the distribution and the important environmental factors. Jill Thompson (CEH) offered to provide some contacts to try to fill these gaps. The site-level data collation has been extended for an additional year as some large datasets from North America and the Southern Hemisphere are expected.
In terms of factors affecting known Phytophthora distributions, the team will be considering host, disturbance by livestock, vehicle movement and landscaping projects, seasonal weather variability, extreme weather events, agricultural and forest management practices, pollutants causing plant stress, soil pH and other edaphic factors as well as non-climatic factors.
Beth and Louise raised the need for data cleaning, for example they need spatial precision (latitudes/longitudes). There is also the issue of species ambiguity, as some species have one or more synonyms or have been re-named since records started. The method used for species detection also needs to be taken into account. For example some records are based on species isolation into pure culture, and others based only on DNA findings. Beth and her team need to know how sensitive and specific each detection method is, whether there have been changes in methods over time, and how reliable is each one?. Beth would like to score the reliability of each diagnostic method. They don’t need to do this for trade modelling, but for niche modelling they do.
In the following discussion it was confirmed that there are not many examples of species complexes now separated, only P. megasperma, P. cryptogea and P. citricola. Records can be distinguished based on whether from isolations into culture, specifies-specific qPCR to detect a unique DNA sequence, standard PCR + sequencing (usually of ITS region), and the recent increased use of metabarcoding (also currently mainly based on the ITS) to detect many species in a given sample. Sarah Green (FR) will outline each diagnostic method and its reliability and pass on to Louise.
The Phytophthora traits’ database was completed in December 2016 for 177 species listing 15 different traits per species (including host range, mating system, survival structures, dispersal mechanisms and thermal tolerance range). It will be updated as new species are described. The modelling team are developing impact metrics using the traits’ database. They will carry out traits-based analyses of Phytophthora impacts to determine which traits influence risk of establishment and spread. Louise ran through some initial results on which trait syndromes best predict the global impacts of Phytophthora species.
Sarah (FR), David (JHI), Beatrice (FR) and Ana (FR) helped identify 24 Phytophthora species present in the UK, and 30 species not present in the UK which the team will use to develop the niche-distribution models.
iii) Horizon scanning
Mike Dunn (FR) spoke about assessing tourism routes, in particular gathering information on visitors to UK parks and gardens. There are 31 million visitors to the UK each year; one third of these visit public parks and gardens. Mike would like to collect more information including where the visitors come from, at what time of year and whether site managers have disease concerns. Mike asked for ideas on how to gather this information, for example from visitor books and tour operators.
The WP3 session finished up with Beth asking for the names of people/organisations who might potentially use the outputs of their work.
14.30-15.00: WP5 Synthesis and integration – Sarah Green
Sarah Green (FR) spoke about WP5 which deals with the overall project coordination, communication and interaction, promoting information exchange and interdisciplinary practice. She talked about WP5 achievements over the last year including establishment of the project data sharing tool (Huddle), the project website, the Expert Advisory Panel and the Project Board, which has met four times over the last year, as well as the two all-project team meetings held during the project’s first year.
WP5 also co-ordinates the Science-Practice-Policy Network (SPPN) involving to date the organisation of a well-attended workshop (~50 attendees) held on improving nursery resilience to Phytophthora (in October 2016), a short article on the project written for the Arboricultural Association Magazine, and attendance (Tim Pettitt [Uni of Worcs] and Jane Barbrook [APHA]) at the National Plant Show in June 2016. Tim also attended the Four Oaks show and other growers’ meetings to raise awareness of this project. Team members also attended international conferences over the last year and gave presentations on the Phyto-threats project.
This year the Phyto-threats team will have a stand at the National Plant Show, held in Birmingham in June. Ideas for the stand and a 15-minute seminar slot are still being developed. Sarah and Mariella will also attend the IUFRO 125th Anniversary Conference to be held in Freiburg in September and give presentations about Phyto-threats. A second stakeholder workshop will be held at FERA, York, on Oct 3-4th this year with the aim of identifying effective management options to underpin an accreditation scheme.
Finally, research result summaries which were produced for the end of year report to THAPBI will be posted on the project website.
Ethics committee.
Mariella Marzano spoke briefly about the ethics committee, set up to review project protocols. She reiterated that in order to use data or photos we need to have written permission from the source. All work packages need to be committed to the ethics values and we need to be consistent in our use of anonymity when sharing data. We all need to think about how we are getting consent for sampling, interviews and surveys, and we need to have clear protocols for retaining and storing data. These issues will be discussed further at the next board meeting.
Tour
The meeting ended with a tour of the JHI pathology lab and sequencing facility.
Phyto-threats project team meeting
October 3rd 2017, APHA, Sand Hutton, York
The aim of this meeting was to bring the entire project team and members of the Expert Advisory Panel together to share and discuss research progress since the last all-project team meeting on May 4th 2017, and to outline and receive feedback on future research plans.
The meeting started with a welcome by Sarah Green (Forest Research, FR) and brief introductions from everyone present including their affiliations.
09.30-11.00: WP1 Phytophthora distribution, diversity and management in UK nursery systems – David Cooke (JHI) and Pete Thorpe (JHI)
David Cooke introduced the WP1 team and reminded everyone of the WP objectives and methods used for the fine-scale sampling of the 15 partner nurseries, including some of the practical issues. A simple questionnaire is applied to each nursery to collect basic data and to get to know the manager, asking for example how material comes onto site and where it goes off site, where material comes from, where it goes to, what water sources are used (ie borehole/river/pond) and how it is treated, and whether the manager has any particular concerns. David explained that sampling involves collecting material from known and unknown Phytophthora hosts, some common to all sites and a mix of symptomatic and asymptomatic. Plant tissue collected is mostly roots. For water sampling, water is passed through batches of potted plants in trays which are left to sit for 30 mins or so. This allows for any Phytophthora propagules on the roots or in the potting soil to be flushed out. The flow-through water is then collected and filtered to trap Phytophthora propagules on the filter. Water supplies and water collection areas (ie puddles, irrigation ponds, drainage ditches) are also tested and filtered. David reminded everyone, and planned to reiterate to stakeholders at the workshop the following day, the importance of water in Phytophthora spread and that water management in nurseries is a critical area to get right in order to control disease. On each nursery site the team evaluates potential disease control points and contamination hazards, but it’s always a balance between time available and the need for detail.
In terms of a sampling update for the fine scale sampling, the WP1 team have carried out 34 sampling visits to the 15 partner nurseries (6 in England, 1 in Wales and 8 in Scotland). Thus each nursery has been sampled at least twice, and a third round of sampling is currently underway, with a total of 1700 nursery samples plus associated meta-data collected so far. Just over 400 samples have been PCR tested for Phytophthora: this includes 93 plant root samples from 35 different host types, 132 water filter samples and 170 samples of buffer associated with the filters. Isolations have also been attempted from selected samples, resulting in three confirmed P. austrocedri findings and a finding of P. cambivora on shelterbelt trees.
For the OPAL community sampling, carried out through co-operation with David Slawson and Vanessa Barber of the OPAL project, the sample kits were sent to the volunteers in June 2017 and a training course/skype video made by David to inform volunteers of the sampling protocol. Ten samples from OPAL have been received to date from various sites in North Wales, all collected by one of the OPAL volunteers. David also showed a leaflet that is being used to explain the project and to aid understanding of Phytophthora diseases.
The broad scale nursery sampling is carried out as part of the national statutory sampling programme in liaison with Alexandra Schlenzig (SASA) and Jane Barbrook (APHA). This element of the project targets 50 nurseries/garden centres in England and Wales and 25 in Scotland (to be sampled in 2017/18). Sample packs were sent out to Plant Health Inspectors earlier in the summer and 27 sample packs have been returned so far (ie representing 27 nurseries; 11 in Scotland and 16 in England and Wales). Each sample pack contains 5-10 different root samples per nursery as well as a limited amount of information on each nursery.
For Phytophthora detection using metabarcoding, the Phytophthora-specific PCR assay is the first key to understanding how many samples are Phytophthora +ve. Currently JHI has a large backlog of samples, and they are splitting the lab processing with Forest Research, who are dealing with the root samples. There are no data yet on Phytophthora species findings as the first Illumina plate failed quality control, probably due to sample loss during one of the clean-up stages in the library preparation. The plate is being redone with the aim of completing the Illumina runs this month. The sequencing runs will also include synthetic control sequences generated as a test of; a) sequencing error, b) indexing error and c) sensitivity range.
The team at JHI have been putting together a database of known, verified Phytophthora ITS sequences, comparing databases from Santi Català, Treena Burgess and a database developed by David Cooke. Phylogenetic trees have been produced for each database so that duplications and variations among sequences can be observed, including sequence errors, for sample one problem has been the truncation of even some type-strain sequences. A new database is being constructed through manual editing of the phylogenetic trees for the existing databases, for example if three sequences for a given species are identical in each database then any of the three sequences can be used, and any sequences with errors are discarded.
David presented some of the slides he plans to show at the stakeholder meeting the following day, including Phytophthora findings by nursery (anonymised), findings by sample type, and observations while sampling. In terms of risk of Phytophthora coming onto site there is the need to keep water sources clean, to be aware of the health and source of plant material coming in (this presents high risk especially if from EU or third countries), and to consider biosecurity for staff and visitors as mud is a problem in some nurseries. He recommended a concrete pad for delivery/despatch areas. In terms of Phytophthora dissemination on site, plant to plant spread tends to be least problematic in cells raised above ground, and since puddles are oftenPhytophthora +ve then drainage is important. Infection by Phytophthora has been picked up in shelterbelt trees and having nursery ‘hospital/recovery’ areas is not a good idea. Rapid disposal of sick plants is optimal. David also recommended quarantining new plant material if possible – this material is often put at the back of the nursery in unkempt areas. David then went on to show photographs of some of the issues encountered during the nursery sampling, ie puddles, muddy and/or flooded ground, soggy possibly Phytophthora-infected roots. Photographs of good management included covered water-holding tanks, collection ponds that are lined and free of vegetation, well-built drainage ditches, graded surfaces that minimise puddling in key parts of the nursery, for example where vehicles move in and out, plants sitting on raised benches, on well drained gravel or clean MyPex with good spacing between plants.
The next steps for the WP team are to complete this autumn round of sampling, accelerate the lab testing, run the metabarcoding analyses to identify the species present, complete the computational biology platform, report species findings to nurseries and begin data interpretation.
Questions and comments:
Q: What about the implications of finding Phytophthora in the nursery – what do they do about it ?.
A: Being aware of symptoms is important and we make management recommendations when reporting on where samples have proven to be positive for Phytophthora. For example if roots from a specific supplier are consistently Phytophthora +ve then avoid that supplier. Also, not all Phytophthoras are pathogens – although we need to be careful when making this statement, for example P. gonapodyides is also a pathogen as well as being fairly ubiquitous in water.
Comment: It is early days yet in the project and as results come in they will help to indicate how nurseries can reduce Phytophthora through making changes to certain practices. In the longer term the project will be able to offer very good advice, very targeted, to guide management.
Q: What about findings of statutory importance ?.
A: Findings based on DNA data alone cannot be a basis for statutory action. The nursery manager however needs to be aware, and Plant Health kept informed.
Q: Is there competition between so called pathogenic and non-pathogen Phytophthora species, and if you get rid of the non-pathogens are the pathogens more likely to take hold ?.
A: Interesting point. A study in Austria is looking at whether some Phytophthora species are outcompeting others, and investigating the interactions among species.
Q: What is the best way of killing Phytophthora in plant disposal areas ?.
A: By composting to a standard that ensures the required temperatures are reached. There are numerous published studies on this. For example Fera protocol on P. ramorum suggests that composting will be effective in killing this species. Any waste disposal system will need to be built into the accreditation system.
Bioinformatics analyses
Pete Thorpe (JHI) presented on the metapy bioinformatics pipeline developed by himself and Leighton Pritchard at JHI. This pipeline is freely available on the GitHub open source site. Pete reminded everyone of what the pipeline does including the five clustering tools. A new clustering tool has been recently published (ZOTU: Exact Sequence Variants: Callahan et al. 2017). ZOTU explicitly tries to correct PCR and sequencing errors and has now been incorporated into metapy. There was some discussion on ZOTUs versus OTUs. Leighton Pritchard explained that ZOTU tries to account for systematic sequencing errors before it clusters sequences. This is different from biovariation, which we are interested in. Pete then showed the mathematical model that ZOTU uses for this correction.
Pete also emphasised the importance of the database as the critical determinant of classification accuracy and he showed the differences in outputs from the five clustering tools, and explained the need to trim the ITS sequences to include the ITS1 region only. BLASTClust is not specific enough and so will be removed from the pipeline. The next steps are to verify the pipeline and database with the control samples from the sequencing plate and to write a Bayesian based clustering/probabilistic model.
Questions and comments:
Comment: In the POnTE project, which compares metabarcoding detection of Phytophthora with a traditional baiting method, sometimes metabarcoding has not found a species when it has been baited out of the same sample!. Something is not right if this happens.
A: Metabarcoding is never going to be 100% accurate, however we are striving to get it better. If a species is not being picked up then this may be because the sequence is not present in the database. The database team are meeting to discuss this.
Comment: One issue has been with false positives, due in some cases to ITS sequence variability within species. Most ITS sequences for species in GenBank are Sanger-generated so will be the most abundant/easily amplified sequence in that species that is deposited as the GenBank reference sequence. Illumina sequencing has such great read depth that it will also generate reads for less abundant ITS sequences in a species. For example, an ITS sequence present in P. gonapodyides also seems to match P. mississippiae. In these cases the less abundant sequence will occur at low read numbers in the presence of higher read numbers of the most abundant sequence. So it can be picked up, though this emphasises the need for data verification/interpretation by those who know the species and their sequences.
A: Yes there will be sequence variants within species – these can be pulled out and identified.
11.00-12.30: WP2 Feasibility analyses and development of ‘best practice’ criteria – Mariella Marzano (FR), Glyn Jones (FERA) and Colin Price (free-lance academic consultant)
Mariella reminded everyone of the WP2 team members, objectives and methods, including the consumer survey (1500 respondents) which explored the plant buying habits of the public (reported on at the last project team meeting on May 4th) and the interviews with nursery managers, of which six have so far been conducted. An online smart survey has also been produced targeting a broader range of consumers including nursery owners, garden centres, landscapers and plant-buying members of the public. This survey is being circulated via a number of online avenues.
For the nursery interviews, carried out by Mariella, Mike Dunn and Tim Pettitt, a range of questions are asked on what influences their decisions, where nurseries are least and most able to change, and their perspectives on accreditation. The aim is to interview all fifteen partner nurseries in the project this financial year. Mariella then ran through a number of slides illustrating some of the comments/perspectives received so far from nursery managers on issues such as plant health, consumers, biosecurity practice/challenges and accreditation. Some points to consider on best practice are that even nurseries who don’t import might unknowingly buy plants from another nursery that does import. Some nurseries aim to be ‘green’ by re-using plastic pots etc, however reducing plastic waste in this way also increases disease risk. In terms of accreditation there is cynicism. Source is an issue, for example a plant that came from Holland and arrived in Scotland – is it fair to say it’s locally sourced ?. Also, landscapers are asking for plants based on design rather than asking nurseries what’s possible and suited. This pressurises nurseries to arrange risky imports. If a nursery cannot import then the customer will just get it elsewhere. Some nursery managers feel that accreditation is just a tick-box exercise; customers don’t ask for it, and there is little support for accreditation at present. The most popular place to buy plants now is at the big retailers and garden centres, and these are viewed as being not so concerned with biosecurity. There also seems to be a perception that accreditation would not change the behaviours of fellow nurseries. However, some nursery managers might consider accreditation if the costs were not prohibitive and the required actions not unreasonable, if there was a safety net and a demand from the consumer.
Mariella then posed a suggested list of questions for the focus session at the stakeholder workshop to be held the following day and the ensuing discussion largely centred around those questions.
Questions and comments:
Comment: Other projects led by Fera are asking similar questions and we’re finding that retailers have less interest in talking about pests and diseases than growers.
A: It’s about figuring out how to talk about pests and diseases. Garden centres and superstores don’t want negative messages.
Comment: At the stakeholder meeting tomorrow we should talk about the consumer survey message on willingness to travel further to support accreditation. There is willingness, and it is important to show this.
Comment: Are we using the wrong language to talk about accreditation ?. It’s not about enforcement or accreditation having ‘teeth’, but rather ‘what’s in it for me ?’. For example, if we get accredited and there is a big government planting scheme, will you buy our plants in preference?. Think of the positive benefits, not what will happen if we don’t become accredited. Perhaps the question tomorrow needs to be phrased ‘what would you like to see from government?’. Could the government make the climate more effective for accreditation?’, ie if the government has a planting scheme would the government only use accredited plants ?.
A: We shouldn’t mention the government specifically, but rather ask ‘what support is needed for accreditation ?’. Then this doesn’t focus on support from a particular sector.
Comment: Use the term ‘credibility’ when talking about accreditation. Don’t underestimate the influence of landscape architects and materials coming straight through Dutch orchards. How do you deal with that ?. Government involvement is a very good point, it is needed for credibility.
Comment: How is the Phyto-threats project addressing existing initiatives for accreditation schemes, for example UK Sourced and Grown and the HTA-led assurance scheme ?. Discussions have been going on with these schemes, you need to know where they have got to.
Comment: We should ask growers how they think they can persuade consumers to buy accredited plants and focus on the positives, ie more healthy, beautiful plants that improve quality for the producer and greater profits too !. What would they need for this to happen ?., what are the economics?. There should also be a clear logo/badge for such plants.
Q: What about a list of possible incentives? What are the scenarios? Best case, Draconian to almost utopian. For example ‘Landscapers like disease-free plants’. Or is it more likely ‘you’re not allowed to sell unless you have accreditation’ (i.e., that accreditation is mandatory)?
A: Accreditation needs to be down the whole line, from landscapers right back to source.
Economic feasibility of best practice and accreditation
Glyn Jones presented on Fera parallel activities including the Forestry Commission decision tool project (a generic decision tool for assessing response options to tree pests in the UK) which is relevant to the Phyto-threats project as it might be applied to new Phytophthora outbreaks in the future. Glyn outlined the tool including why it was needed, what it does, how it was developed and its limitations. A draft final version of the tool is now in review. Essentially, the tool was developed because of the need to provide support on the economic impact of new or unknown species in a new area within a very short timeframe. The idea was to produce a single model for all pests and diseases for all end users using a standardised framework for scenario assessment. Development of the tool required input from a wide range of end users (economists, scientists, policymakers) who formed a steering group.
Glyn ran through the model and its development in his presentation. It is a combination of a prevalence model (based on pest/disease surveillance and a rule of thumb on how hard you are looking versus the actual infested areas found plus an epidemiological model) linked to an economic impact model. Glyn listed at the end some of the model’s limitations including the spread model which assumes a constant spread of the pest/disease over time. Also there are very few control options for new pests and diseases, so not being able to input various control options into the model is a weakness. There are also uncertainties as to which environmental values to incorporate into the model. Glyn finished this part of his talk with the questions; will the model be used ?, should it be used ?. It is a highly generic model that can be abused. How should it be used ?.
Finally, Glyn outlined plans for a workshop to be held in November 2017 on cost and responsibility sharing by industry when pest and disease outbreaks occur. Points to be discussed at the workshop include current biosecurity activities, how an industry scheme might work, and incentives to join. There will be cross-reference with the Phyto-threats project via Mariella and Gregory.
Questions and comments:
Q: Can you put case studies in, for example P. ramorum, horse chestnut bleeding canker ?.
A: Yes, and they are there.
Q: Is this decision support only for outbreaks ?. Is there decision support for interceptions ?.
A: Probably this tool could be adapted for interceptions if it were represented as the minimum area for an outbreak.
Colin Price then gave a presentation demonstrating how an adapted version of the CARBBROD model, developed for Dothistroma, could be used in a scenario analysis to estimate the cost of potential timber and carbon impacts when Phytophthora outbreaks occur. This analysis will be done in year three of the project to explore the wider costs associated with no change to UK nursery practice (i.e. a continuation of the status quo in which new Phytophthora impacts can be expected) compared with a scenario where changes to nursery practice will reduce the likelihood of future outbreaks by new Phytophthora species.
The model is based on Forestry Commission yield models. It uses DECC (Department of Energy and Climate Change) carbon prices based on international agreed limits, or the social cost of carbon (i.e. the effect of increasing CO2 concentration), or any other carbon price schedule. Discounting is done using the Treasury’s schedule of discount rates, or any other schedule, or a single discount rate. When there isn’t a unique carbon price, and when discount rates vary, every year has to be evaluated on its own, and all individual values summed. Thus tracking carbon pools into the indefinite future is a challenge. Colin showed how the model predicts the optimal rotation value for Japanese larch. Sometimes CARBBROD gives better carbon values when there is infection in trees, using the DECC values. This isn’t the answer we would expect. Other carbon prices give more expected results.
Questions and comments:
Q: Has CARBBROD been tested against real-world data? i.e. documented case studies? Do the predictions emulate what really happened?
A: All data come from the real world!
Q: Can you add more than just carbon values to give you a more intuitively correct answer?
A: Various impacts of trees have been looked at, for example temperature reduction, pollution absorption, flood alleviation. Carbon values are so big that they will always dominate models.
Comment: More talks with pathologists are needed since it is not clear what the primary inputs would be for the Phyto-threats project. Assuming best nursery practice, what scenarios can we predict? This is a two-way discussion between economists and pathologists on the team and we need to identify over the next six months specific scenarios of use/interest to the project that Colin can apply to CARBBROD. For example WP3 might identify specific high-risk Phytophthoras not yet present in the UK, but which might enter the country if no change in existing practice occurs, that could be applied to CARBBROD.
A: Gregory/Colin will initiate these discussions by email.
Team meeting afternoon session
13.00-14.30: WP3 Global Phytophthora risks to the UK – Louise Barwell (CEH)
Louise began with a run through of the WP3 team members, objectives and associated milestones. For objective 3.1 (Risk of Introduction) a year 1 milestone has been to compile a global country level database of Phytophthora occurrence. The team has now amassed 13,853 country-level records covering 1601 Phytophthora species x country combinations including year of first record of that species in the country and invasion status. The team are aware of potential sources of bias in country-level occurrences such as level of pest recording activity per country, biosecurity, and the lag phase between pest arrival and first report. Louise has collected metrics on national pest reporting activity and posed the following question to the team; how can national recording and biosecurity efforts best be measured, ie are there descriptors of Plant Health Inspector activity ? Can we weight records against the number of inspections per country ?
Currently Louise is looking at assessing national pest reporting activity through the IPPC, legal instruments such as ECOLEX and level of biosecurity investment (from the FAO). The team have been doing a preliminary analysis developing the first models based on post-2000 occurrence data using predictors such as trade connectivity, recording effort and biosecurity. Live plant imports on their own explained 44.36% of variation in Phytophthora arrivals per country since 2000. When combined with metrics of recording effort per country, the model explained nearly 60% of the variation in Phytophthora arrivals. There followed some discussion on the use of the term ‘arrivals’ vs ‘records’. Some of the post-2000 increases in Phytophthora records per country might not necessarily relate to new arrivals, but rather to increased surveillance effort (ie expansion of molecular tools) reporting species that may have been present in that country for some time. David Cooke suggested weighting the new records according to species description date.
The team have also looked at trade pathways and tried to link Phytophthora reports in each country with different commodity imports. The resource trade data allow for live plant imports to be broken down into different commodity types, so it should be possible to get a sense of which types of hosts seem to be transporting the most Phytophthoras. The next steps are to collate further trade networks data, to break down total Phytophthora ‘arrivals’ to the species level, and build in country by country climate matching metrics. Once species-level arrivals can be used as the response variable, particular traits can be assessed for whether they make species more or less likely to arrive. The final output will be to predict the arrival risk of different Phytophthora species to the UK and to simulate how this might change under different kinds of trade scenarios. So data will be sought on different projections for future global trade.
For objective 3.2 (Risk of Establishment and Spread), the team have so far collated 9907 georeferenced global Phytophthora records with 1-10 km precision. The data encompass 81 species from 38 countries. In collating these data the team are prioritising regions that are climatically similar to the UK. Louise showed a slide listing the data sources and highlighted the sources of uncertainty (ie taxonomic uncertainty due to differences in use of different ID methods, positional uncertainty, recording effort resulting in false absences etc) and how such uncertainties might be overcome.
As part of objective 3.2 the team are developing niche models for different Phytophthora species predicting global impact and risk of establishment and spread in the UK. As an example, Louise showed the global distribution model for P. cinnamomi published by Burgess et al. 2016 (in Global Change Biology) and presented a list of the potential environmental drivers of Phytophthora distributions ie animal activity, pollutants, hot and cold stress. The pathologists on the Phyto-threats project sent Louise a list of 54 focal Phytophthora species with known or potential impacts on UK tree species for the niche models, including 24 species currently known to occur in the UK – this list can be found on Huddle. These species will be used to validate how well the potential distribution of Phytophthora species in the UK can be predicted with the global niche models. The other 30 species are absent from the UK and 15 of these are thought to also be absent from Europe. For these species risk maps will be created, and each one will be ranked for its potential impact in the UK.
Louise also described the global Phytophthora traits’ database, part of which has now been merged with a similar traits’ database developed by a team based in Australia and New Zealand. As a result of this merging of databases we now have a comprehensive traits’ database complete for 172 Phytophthora species which it is thought could be managed and updated by Scion (New Zealand) over the next 10 years as part of their long-term Phytophthora research programme. The two groups aim to publish the traits’ database in the near future, after which it will be made freely available online. Louise then ran through some of the traits included in the merged database (i.e., characteristics of sporangia, oospores and other survival structures, thermal tolerances). Not yet included in the merged database are data on species distribution, host species, genome size, disease symptoms and impact metrics. These data will be added later following the publication of some manuscripts currently in preparation.
Louise ran through some of the questions they are testing using the traits’ database. These include examining the value of traits’ data for pathogen groups, the extent to which traits’ databases been compiled for pathogen groups in the past and whether these traits can be used in analyses of pathogen behaviour. Also, is there a phylogenetic signal in the traits ?, ie to what extent do closely related species share traits, which traits seem to be experiencing the strongest selection pressures, and are there groups of traits that are co-evolving in response to global movement of Phytophthora ?. Another question is, do hybrids share traits with parent taxa?
Louise then showed a Phytophthora phylogeny based on the ITS region that had been sent to her by Treena Burgess in Australia. Louise has had a first attempt at measuring the phylogenetic signal in some traits, with the outcome being that there is generally a lack of phylogenetic signal in traits. Only sterile and non-caducous species had a non-random relationship, and there was a nice correlation between trait and cumulative branch distance between two species. This analysis used a ‘Brownian motion’ model of traits applied by ‘crawling’ down branches and flipping at random. Louise asked the project team what other appropriate models could be used for trait evolution for Phytophthora ?. The response was that the project team aren’t familiar with the different models, but David Cooke suggested focusing on nodes as that is where species split. If Louise could circulate some of the models that she has in mind then others will be able to advise. It was also noted by members of the project team that a multigene phylogeny might be better for the analyses (i.e., Martin et al., 2014), although the ITS-only phylogeny would probably not differ that much. David Cooke commented that it would also be nice to relate genome evolution with the trait database.
The other question being addressed is, can species traits predict or explain the global impact of Phytophthora species? That is, what makes an invasive species? Two impact metrics, geographic extent and host range, are being used to test this by modelling individual traits vs groups of traits (‘trait syndromes’); the idea being that particular combinations of traits will help a species get all the way along the invasion pathway (introduction > establishment > spread > impact). Louise has looked at trait syndromes and the extent to which certain traits might be positively or negatively correlated. For example being caducous and causing foliar disease are positively correlated, whereas being sterile and causing root disease are negatively correlated. Heterothallism and invasive potential on roots (in particular) and foliage are key traits that co-vary and are positively correlated. It could be anticipated that heterothallism allows greater sexual recombination and adaptation.
In terms of whether trait syndromes can explain global impacts, initial results suggest that high impact species (ie those with the broadest host ranges) tend to be heterothallic, have one or more survival structures, broad thermal tolerances and fast growth rates. Host range can also be predicted by root (and foliar) disease symptoms. In general, trait syndromes are more ecologically informative about the global impact of Phytophthoras. Louise went on to suggest that it might be possible to develop a trait-based early warning signal for Phytophthora pathogens which are known to have certain traits, but for which there are no data on impact, ie to try and predict the behaviour of a novel species at an early stage. In this way potential threats can be ranked in order to inform Pest Risk Assessments.
In terms of next steps, the WP3 team will publish the traits’ database and phylogenetic analyses, and they aim to submit a paper linking traits and global impacts this November. Louise will continue to collate data on global Phytophthora occurrences, fine tune niche models and begin preliminary niche modelling approaches. They aim to co-develop final model outputs with policymakers.
14.30-15.00: WP4 Predicting risk via analysis of Phytophthora genome evolution – Ewan Mollison
Sarah Green introduced Ewan Mollison who recently started (August 1st 2017) on the project as a PDRA working with Paul Sharp at the University of Edinburgh. Ewan started his presentation with a project overview and talked about the different mechanisms of evolution in oomycetes ie vertical gene transfer, horizontal gene transfer and hybridisation. Horizontal gene transfer, which is the direct transfer of genes between two species rather than from parent to offspring (known as vertical gene transfer), is of interest because oomycetes are known to have acquired genes from fungi in this manner. Ewan is interested in knowing to what extent this occurs in Phytophthora. Such events can be traced in genomes since ‘recently’ acquired genes from another species may look quite different to neighbouring genes as they tend to retain the characteristics of their donor species. Hybridisation events can also be identified in genomes by increased genome size and chromosome number or by analysis of gene sequences. Ewan presented a slide illustrating hyphal anastomosis and zoospore fusion; two possible mechanisms by which horizontal gene transfer and hybridisation may occur in Phytophthora.
The key aims of the project are (in the shorter term, ie the next six months) to sequence and assemble the genomes of three Phytophthora species that appear less damaging, but are closely related to species highly damaging to woody hosts, and over the next two years to characterise key genes involved in woody host vs non-woody host infectivity, and to look for signatures of horizontal gene transfer and hybridisation between species.
Ewan presented a brief overview of Phytophthora genomes with the most extensively studied being P. infestans, P. ramorum and P. sojae. Genome sizes are variable, for example the P. austrocedri genome is 140 Mbp and consists of 47% repetitive regions. All species with annotated genome data have from 14,000 to 18,000 genes. Ewan then showed where all the current 25 genome-sequenced Phytophthora species sit by clade using the cox2 gene to calculate the trees. He went on to describe the characteristics and phylogenetic positioning of the three species to be target-sequenced in this project. In terms of looking at woody vs non-woody host infecting species, Ewan will look at ways in which their gene complements differ, whether characteristic sets of ‘woody host infecting’ genes can be identified. He will start with a comparison of P. ramorum (woody) and P. sojae (non-woody) as these are the most extensively studied species. Ewan then finished up with an overview of some background work on genome sequences of P. austrocedri isolates from Britain and Argentina, looking at similarities among isolates and between isolate groups, and whether the genome sequences might tell us anything about the origins of the two outbreaks in each country.
Questions and comments:
Q: How much of the difference between Argentinian and British isolates occurred in the source population before their separate introductions into their new regions ?.
A: We have no idea of the size (ie genetic variability) of the source population and can only infer what has happened since the founder population began in each country. In this case spread has been clonal, so the differences between the two isolate groups have largely occurred in the source population.
Q: It wouldn’t be surprising if these invasions were driven by multiple introductions. Is it possible to look at this ?.
A: Yes this scenario is likely, although the separate introductions were likely to have been of the same clonal strain in each country.
15.30-16.00: WP5 Synthesis and integration – Sarah Green
Sarah Green gave an overview of WP5 activities since May 2017, including Board meetings and the reports/research summaries recently posted on the project website. Sarah noted that a four month no-cost extension (ie to July 31st 2019) which applies to the whole project and its work packages has been agreed by the funders. Jill Thompson (THAPBI co-ordinator) followed up on this point where it was confirmed that the whole group can spend existing funds up to the new deadline (but the project will not be in receipt of any additional funding). Project reporting will be three months after the new deadline. Sarah will initiate changes to the Collaborative Agreement to reflect this.
There was some discussion of where and when to hold the next all project team meeting. It was agreed that CEH in Wallingford next April/May would be appropriate and Debbie will circulate a Doodle Poll to confirm a date once Louise has confirmed the plan with Beth. Sarah then reported briefly on the Phyto-threat’s attendance at the National Plant Show back in June 2017, and talked through the seven posters that were designed for the stand display with the aim of raising awareness of the link between Phytophthora outbreaks on trees in the wider environment and spread of these pathogens in trade. It is planned that the project team attend the National Plant Show again in 2018 with some information presented on project results. Sarah also reported briefly on the 125 thIUFRO Anniversary Congress held recently in Germany, where the Phyto-threats project was represented by a talk and poster. Jill Thompson also mentioned an event to be held at Dynamic Earth in Edinburgh next month, it will be mostly targeting school children, but the project could have a couple of posters there. The next stakeholder event to consider will be the THABPI dissemination event to be held in London on February 7th 2018. Watch this space!
Phyto-threats workshop
Improving nursery resilience against threats from Phytophthora
6th October 2016, APHA, Sand Hutton, York
Sarah Green, Phyto-threats project co-ordinator, welcomed everyone and introduced the aims of the workshop. The aims of this first workshop were:
- to introduce the scientific aims of the project
- to develop collaborative networks across individuals and groups with an interest in working towards collective best practice in nurseries
- to share lessons and experiences around the challenges and opportunities of managing disease threats
- to identify nurseries and other stakeholder groups and individuals who could and would become involved in the research
The meeting was attended by c45 academics, nursery managers, Plant Health inspectors, foresters, policymakers and others
Richard McIntosh (Defra) provided a policy context for plant health in the UK. He emphasised the value of healthy landscapes and the benefits they can provide. Increased vigilance is important, but also having the capacity to quickly intervene in the event of an incursion. Richard outlined the 5 ‘Ps’ approach (predict, prevent, protect, prepare, partnering) and highlighted the range of approaches the government is taking pre-border, at the border and inland. He posed a challenge to the audience to think of how they could incorporate a 5’Ps’ approach to their business.
Mike Harvey (Maelor Forest Nurseries) highlighted the changes they have seen in their nursery over the last 20 years, particularly in the numbers of new pests and diseases and he noted the limited range of tools that nurseries have to deal with damage and control. Maelor have invested in Integrated Pest Management which guides use of water, clean areas and purchasing behaviour.
Ian Nelson (Johnsons of Whixley) presented a trade perspective and noted the complexity of the trade network. Business is largely driven by price and profit. He said that UK growers currently cannot meet the UK demand for plants and therefore import from abroad. However, he recognised that the inspection regime in other countries may not be as robust as in the UK. Ian called for greater education among customers such as landscape contractors to look for alternatives to plants that can host serious diseases
David Edwards (Tilhill Forestry) started by describing the devastating impact that Phytophthora ramorum has had on larch forests in South Wales. David explored the efficacy of different approaches to deal with P. ramorum, but also the huge challenges of trying to predict the next outbreak. He made a plea for prevention rather than cure for tree health and warned that any solutions should not impact heavily on the economics of forestry (e.g., abandoning Sitka spruce).
A panel discussion chaired by Sarah Green (FR) elaborated on some of these challenges, highlighting the dilemmas of ‘unknown unknown’ as well as ‘known unknown’ harmful organisms. The use of correct tools for detection was said to be important and discussions touched on the closure of high risk pathways. The value that plants add to the environment is believed to be highly under-valued by society, which facilitates the desire for cheaper products and imports. Reducing bureaucracy was considered to be key to making changes in the sector as well as seeking opportunities to increase the quality and quantity of UK plant production.
Each of the 4 project Work Packages presented a 5-minute introduction to their work.
Work Package 1 (presented by David Cooke from James Hutton Institute) focusses on understanding Phytophthora distribution, diversity and management in the UK nursery system. The team are developing a diagnostic system that can detect Phytophthoras and identify individual species using DNA methods. The team have been collecting samples at a number of nurseries. David thanked the nurseries who have volunteered so far and welcomed more participants.
Work Package 2 (presented by Mike Dunn from Forest Research) involves social science and economics. The core focus on WP2 is a feasibility analysis and development of ‘best practice’ criteria. The research will involve exploration of nursery practices and issues they deal with on a daily basis as well as attitudes towards best practice guidance and accreditation. A consumer survey will be undertaken to understand better plant purchasing behaviours and public attitudes towards accreditation and what this could entail.
Work Package 3 (presented by Bethan Purse from Centre for Ecology and Hydrology) involves modelling global Phytophthora risks to the UK. The team are mapping trade pathways from source countries and ecological zones and linking ecological traits of all known Phytophthora species globally to likely impacts if they were to arrive in the UK. They are looking to learn lessons from past introductions in order to develop a predictive tool.
Work Package 4 (presented by Sarah Green on behalf of Paul Sharp from the University of Edinburgh) will look at predicting risk via analysis of Phytophthora genome evolution. Questions that this work package will explore include how Phytophthoras have evolved to kill trees, why they are so adaptable and how they can hybridise. The team will sequence the genomes of three Phytophthora species which will add to a comparative analysis of genomes across a range of Phytophthoras.
Two international speakers provided an overseas perspective on Phytophthora risks and nursery accreditation. Susan Frankel (US Forest Service) focussed on California and the impact of Phytophthoras on native plants and wildlands. She noted the unintended consequences of restoration projects that are introducing Phytophthoras. The US National Plant Board have started a certification scheme with 8 nurseries currently signed up (pilot phase). Susan emphasised that complying with the certification standards required a lot of work, but the nurseries involved are then free from intensive inspection per pest. There is also a voluntary accreditation scheme that involves following best practice guidance. Susan recognised that the scheme was not easy and she said you couldn’t claim to be Phytophthora-free, but you could claim to have done everything possible to be disease-free following a systems approach. Giles Hardy (Murdoch University, Perth) was unable to attend in person, but presented a video on the Australian NIASA scheme whereby production nurseries and those involved in growing media sign up to follow best practice management guidelines. This scheme has been in operation since 1997 and the guidelines are now in their fifth edition. Giles described the guidance in detail including crop hygiene, crop management practices, general site management and water management. Giles also described in detail a method for composting. Alongside the nursery accreditation guidelines there is also a national nursery and garden industry biosecurity plan which focuses on risk mitigation.
There followed a workshop session led by Mariella Marzano (Forest Research) to explore what a nursery accreditation scheme would look like in the UK. Feedback suggested that accreditation might need to be tailored for different stakeholders, but possibly under a single umbrella. A scheme could include different levels of standards to encourage businesses to improve their practices. A number of practicality issues were raised and need further exploration. However, there was a strong consensus that any scheme should have minimal bureaucracy. It was felt that there would need to be consumer support for any scheme to provide an incentive for nurseries to be involved. Decisions over what the scheme should include would best be made by representatives from a mix of sectors. Brexit might provide an opportunity for the UK to promote its own best management practices and to have more control over quality of imports. There are practices (e.g. mail orders, garden shows, illegal trade in plants) that could undermine an accreditation scheme. The scheme would require consumers to be informed and supportive.
Jon Knight gave a keynote listener talk, reflecting on the day’s discussions. He emphasised that we need to understand market constraints and explore how to ensure that regulations and legislation work better for the sector and consumers. He noted that capacity will have to be increased if we are to produce more ‘home-grown’ plants, but businesses need to be profitable in order to keep trading and that currently involves importing from abroad. He highlighted that if there is a desire to change trading practices then consumers need to be willing to pay more for ‘home-grown’ plants and that involves recognising the value e.g. of a disease-free environment. He made a plea for models to provide some foresight on future risks to facilitate traders becoming more resilient.
Phyto-threats biannual all project team meeting
5 October 2016, APHA, Sand Sutton, York
This event provided an opportunity for the project team to meet in its entirety, including new team members Peter Thorpe (JHI), Louise Barwell (CEH) and Debbie Frederickson-Matika (FR), together with an overseas member of the Expert Advisory Panel (EAP) Susan Frankel (US Forest Service), and other UK-based EAP members David Slawson (OPAL), John Morgan (FC), Richard MacIntosh (Defra) and Kelvin Hughes (APHA). The objectives of the meeting were to share research updates for each workpackage (WP) in the morning, and to promote team-wide understanding of the plant trade and nursery sampling challenges through a visit to Johnsons of Whixley nursery in the afternoon.
WP1 Presentation – David Cooke (JHI) and Leighton Pritchard (JHI)
David Cooke outlined WP1 objectives as using metabarcoding to analyse Phytophthora community structure in nurseries and associated ecosystems with the aim of informing disease management and best practice and to model Phytophthora communities. David reviewed WP1 sampling methods and reiterated the need to extend networks for nursery sampling. David is involved in a number of related projects that can feed into this one, including a THAPBI Phase 2 project on Early Detection and a Scottish Government funded project looking at Phytophthora diversity in different ecosystems in Scotland. Data on Phytophthora diversity in a range of wider ecosystems can be modelled together with data on nursery Phytophthora diversity. David then gave an overview of the metabarcoding approach to be used from sampling in nurseries through to analyses of Phytophthoras present in each sample.
WP2 Presentation – Mariella Marzano (FR) and Gregory Valatin (FR)
Mariella started her presentation with a review of work planned for the social and economic research. This package will cover the feasibility analyses and development of ‘best practice’ criteria for a nursery accreditation scheme. There are three key parts: social-the applicability of best practice criteria; cost-benefit analysis; the development of best practice criteria to underpin guidelines for an accreditation scheme.
In general the project team need to be aware of all the other initiatives in terms of accreditation schemes, for example Richard MacIntosh (Defra) mentioned the Woodland Trust Accreditation Scheme.
Gregory Valatin asked what the expected uptake of an accreditation scheme might be, pointing out that if low the overall impact would be small.
There was also a short discussion on a 2016 paper by Whittet et al. in Land Use Policy journal “Supplying trees in an era of environmental uncertainty: Identifying challenges faced by the forest nursery sector in Great Britain”
This study involved a survey of forest nurseries in Britain in relation to supply of locally sourced seed and domestically produced planting stock for native woodland and hedging markets. It will be important to follow up on which nurseries were surveyed to avoid overlap.
The discussion ended with the comment that consumers’ willingness to pay will be affected by the availability of cheap plant imports.
WP3 Presentation – Beth Purse (CEH), Dan Chapman (CEH) and Louise Barwell (CEH)
Beth outlined the approaches for WP3 objectives 1) assessing risk of introduction of Phytophthoras to the UK via trade and recreational spread, 2) risk of establishment and spread following arrival, and 3) scoping knowledge gaps to try to predict likely future introductions. The WP team will identify and rank Phytophthora threats to the UK and link invasiveness to different traits.
WP4 Presentation – Sarah Green (FR) and
Sarah (standing in for Paul Sharp, University of Edinburgh, who was unable to attend the meeting) gave a brief overview of the objectives of WP4 which aims to predict risk via analysis of Phytophthora genome evolution. This will enable a better understanding of the genetic mechanisms by which Phytophthoras can infect woody hosts, adapt to new hosts, and the extent to which they can acquire new genes through hybridisations or horizontal gene transfer. WP4 does not start until April 2017, however the project has funding to target sequence three Phytophthora species (to be completed this year) and the focus of Sarah’s presentation was how to select these species.
13.30-17:00: Visit to Johnsons of Whixley plant nursery
After lunch the project team hopped into a minibus driven by David Cooke for the 30-minute trip to Johnsons of Whixley, one of the largest wholesale plant nurseries in the country, supplying mainly the amenity sector. The team was hosted for the afternoon by Ian Nelson, production manager, who provided a lively, honest and energetic tour of facilities and account of business practices as well as responding to the many questions asked by the project team. This was a very useful experience for all members of the project team, particularly those not directly working with nurseries, who finished the visit with a much greater understanding of what is driving the trade and associated management practices.
Phyto-threats start-up meeting April 21st 2016
The aim of this meeting was to bring the whole project team together with a range of stakeholders to present the workpackage (WP) objectives, research approaches and programmes of work in order to generate shared understanding, discussion, commentary and advice.
Project introduction presentation (Sarah Green, Forest Research)
NB: presentation links will open in a new window
WP Presentations
David Cooke (James Hutton Institute, JHI) outlined plans for WP1: Phytophthora diversity, distribution and management in UK nursery systems.
Objective 1 of the WP is to use metabarcoding to analyse Phytophthora community structure in different nursery management systems and Objective 2 is a Phytophthora community modelling analysis. David outlined the proposed methods for sampling, with a brief account of sampling theory and bioinformatics and pointed out potential challenges and technical issues that need to be considered. David also gave a quick account of Phytophthora barcoding literature; for example a recent study of four Scottish streams found the DNA signals of 45 ‘species’ of Phytophthora. This emphasised the need for a baseline of the ‘background’ level of Phytophthoras present in the wider UK environment.
Mariella Marzano (FR) presented the work plan for WP2: Feasibility analyses and development of ‘best practice’ criteria.
This work is split into three parts:
- a social analysis of nursery best practice
- a cost-benefit analysis of best practice
- developing best practice criteria to underpin guidelines for accreditation.
Important to the research will be effective stakeholder mapping and understanding existing values, experiences and practices, and attitudes towards accreditation through a minimum of 20 interviews (of different stakeholders) per year. The cost-benefit analysis will involve nursery and consumer surveys to assess cost of implementation of different disease management measures and willingness to pay for accredited stock. There will be exploratory scaling up of survey values to a national level. The analysis will also enumerate the impacts of failure to adopt best practices. An Ethics Committee has been established to review the social science methods and a first meeting (a few weeks ago) has approved the approaches to be used. This committee will reconvene every six months. Anonymization of data will be crucial.
Beth Purse and Dan Chapman (CEH) presented an overview of the programme of work for WP3; Global Phytophthora risks to the UK.
The presentation was split into three parts as follows:
WP 3.1 Trade pathways and risks of introduction: Dan Chapman (CEH) has been working with EPPO on plant pest pathways and predictions of high risk pathways. He presented on connectivity networks between countries based on trade – import vs export matrix and the link with climate similarities and Phytophthora presence/absence data. GDP of a country in the network is also important (as a proxy for effort into biosecurity). The best model uses climate-weighted connectivity through multiple pathways. Host breadth increases invasiveness of pests and pathogens in general – but in this project this will be related specifically to Phytophthora Trade pathways will be ranked, linked to ecological traits of Phytophthora and risk of a pathogen being introduced modelled based on position in transport networks and source intersection. This project will refine the temporal resolution of arrival and spread, incorporating air transport and more pathogen traits in the analysis.
WP 3.2 Risk of establishment and spread: This work will identify Phytophthora spp. with the greatest capacity for establishment and spread under UK conditions. Models range from statistical inferences on observations to detailed models based on organism traits. However pathogen spread varies with invasion stage/extent and pathogen biology might not be well known. The project needs good global incidence data on Phytophthora species from other sources to make more detailed niche maps. Pathogen niches in the UK will be mapped and best-performing modelling methods applied to 40 focal Phytophthora species to predict invasiveness a) can do this by overlapping information on environment in one country compared to that in other countries, b) for more detailed mapping can use Phytophthora biological trait data and specific modelling against climate. Survival traits are also important, ie chlamydospores vs oospores. David Cooke (JHI) commented that dead wood is not a substrate for Phytophthora survival. Ten focal species will be identified for the modelling (from the UK Plant Health Risk Register). After validation with these species a further 25-30 species outside Europe will be identified for application. Pathogens from agricultural crops will be excluded. Data will be sourced from EPPO, CABI, GBIF, DAISIE and PhytophthoraDB.
WP 3.3 Horizon scanning for emerging pathogens – scoping knowledge gaps: Mariella Marzano presented this section, the aim of which is to understand patterns of human movement and how pathogens are transferred to the UK. The focus will be on tourism and other recreational pathways. Mariella raised the question of how to find out who is coming to the UK for recreational purposes and what could they be bringing in terms of plant/soil material?. This work needs data on person and plant movement. Could the project use data from border security?. Priority should be given to known Phytophthora source regions. David Cooke (JHI) commented that visitor books in guest houses might be a useful source of information.
The potential policy impacts of WP3 include contributions to the UK Plant Health Risk Register, global ecological trait databases, publication of habitat/climate suitability maps for pathogens.
Paul Sharp (UoE) and Leighton Pritchard (JHI) presented the overview of WP4: Predicting risk by analysis of Phytophthora genome evolution.
This WP will start in April 2017 and will run for two years. Paul provided a general introduction to molecular genetics using data from a range of organisms to explain how DNA sequences can yield useful information on evolutionary processes leading to (for example) woody host adaptation, including the role of horizontal gene transfer. Paul also provided an overview of complications in DNA analyses due to hybridisation and horizontal gene transfer. This project will look at genes gained and lost across the Phytophthora population using approaches similar to those used to analyse genes associated with infection of woody hosts in Pseudomonas syringae. Leighton Pritchard presented on currently available Phytophthora genome data, describing the usefulness of different genome databases. Most Phytophthora genomes are in the size range of 30-50 Mb, although P. infestans genome is 130 Mb.
Sarah Green presented on WP5, the integration and communication platform for the project.
This is a network to promote information exchange and interdisciplinary practice within the project team. The project uses Huddle to share information and for project/task management. The project board will meet monthly or every two months by phone and the whole project team will meet twice a year. There will also be annual Science-Policy-Practitioner Network (SPPN) workshops involving project scientists, industry and consumer representatives, policymakers, and other interest groups. This year’s SPPN workshop will focus on scene setting and building relationships. The one to be held in the final year will focus on scoping the further development of an accreditation scheme (the goal of the project).
Stakeholder perspectives
Stakeholder perspectives were given by plant nursery participants Alan Harrison (Forestry Commission), Alice Snowden (Cheviot Trees) and Rodney Shearer (Alba Trees).
Alan Harrison is head of the forest tree and seed supply for the Forestry Commission’s (FC) National Forest Estate plantings. He manages three forest nurseries at Newton, Wykeham and Delamere, growing 23 million trees, buying in 5.6 million. The FC does not buy trees from outside the UK, but some of the suppliers may do. Overall they supply ~ 12K Ha planting each year. The main species is Sitka spruce which is grown in the ground or in cells in contact with soil. Approximately 12 % of the stock are broadleaf spp. grown as bare root and in cells. Alan commented that they have much greater species diversity in their stock compared with 10 years ago, mainly due to the wish to diversify forests and in response to climate change forecasts. Lodgepole pine is back in favour, for example, and some of the new species are Taxus, Tsuga, Thuja, Chamaecyparis, Eucalyptus, Cedrus, Juniperus. Some of these are ‘newcomers’ and we know less about how they will behave in Britain.
Some of the issues raised by Alan included importation – are visual checks sufficient?. Should we quarantine them?. Also, do their existing nursery practices (ie growing directly in soil) make infection more or less likely?. Essentially the nursery wants a better appreciation of risk and what they can do about it. For example what are the risks of further host jumps in Phytophthora?, what is the risk of mutation causing increased virulence?.
Alice Snowdon gave a run through of Cheviot Trees production systems with photos. They are a forest nursery, growing cell-grown stock in polytunnels with approximately 25% of stock going to FC under contract. Broadleaves go to FC and some to foresters under grants. They also grow some Christmas trees. The stock is sold at 1-2 yrs old, mostly 1yr old. All stock is raised above ground over MyPex with mist irrigation indoors. The water source is borehole including one from gravel under a river bed. Water drains from beneath/edge of beds. Trays are always washed after use in cold water (this water would be a good sampling point), however, the nursery is considering changing this and would like to know whether it is worth the effort.
Newly sown crops can suffer from damping off. They have also had cases of patch dying of conifers, hawthorn and privet. Samples of diseased stock get sent to a laboratory for testing. Generally the lab reports show many species of potential pathogen organisms – they cannot tell the nursery which is primary. Diseased saleable plants with blackened stems and top wilt are nearly always diagnosed as Phytophthora. This is normally addressed through changes to irrigation. Sometimes conifers are found to be positive for Phytophthora – and treated with fungicides. Impact of Phytophthora currently means changes in species stocked (ie they no longer grow larch and are cautious about Juniper).
The nursery considers that involvement in this study could be risky, but they hope not. They are looking for guidance on best practice. Alice was interested to know more about the Australian nursery accreditation scheme and how it works. In terms of management practice, Alice wondered if the project could test irrigation water sitting in tanks in winter for Phytophthoras. Alice also wondered whether cell density in trays was important in terms of increasing ventilation (less humidity for infections) and how long to allow tree collars to dry between watering to lower risk of infections. Apparently there are very few fungicide options on the market. A phosphite-based compound was reportedly very effective, but was taken off the market.
Rodney Shearer presented on Alba Trees, who grow 11 million cell-grown trees (no bare root). The nursery doesn’t buy in anything and nothing comes from the EU. They have recently acquired a tree nursery in the Czech Republic as an export agent. In Scotland the nursery has two sites about 800 m apart. One site has greenhouses and propagation, and the second site is a farm which does the growing-on. The nursery has the potential to produce 14 million trees if the market was there. Red-band needle blight has reduced the pine stock requirement; apparently there are further restrictions on movement of pine in Britain due to the presence of a unique southern strain of the pathogen not present in Scotland. The nursery has reverted to using disposable trays for susceptible crops because of disease risk, but this causes much plastic waste. They do not use compost, as tends to be from municipal waste and is not uniform. Instead they use a peat based non-sterile product from peat bogs which is tested for Phytophthoras and eelworm. Rodney did express concern about the application of notifiable diseases and exclusion zones, citing an experience the nursery had with fireblight on hawthorn. Alba Trees used to grow mainly native species, but now also stock more alternative conifers. Rodney cautioned on the risks involved as we don’t understand enough about their biology. Alba are not scared to have project scientists visit as they want to know what Phytophthoras they have in order to reduce risk. The nursery wants stability and they need to know the balance of risk and species.
Policy and industry perspective
Members of the Expert Advisory Panel gave their talks from policy/industry perspectives.
Kelvin Hughes, Chief Plant Health and Seeds Inspector, APHA;
APHA have 190 staff and cover 30 UK border inspection points. There is 18h/day, 365 days/yr cover at major points of entry. They do passenger baggage checks too and general surveillance. APHA do Plant Health diagnosis with R&D done by FERA. Traditional techniques for diagnosis dominate, although they are now promoting the use of on-site molecular diagnostic instruments (ie Genie machines) for tests applicable to specific pathogens.
The UK is responsible for 1/3 of EU notifications and the current main Plant Health issues are Epitrix and Xylella. Kelvin stressed the importance of collecting plant passporting information during project sampling. It is important that the project works with PHSI in sampling, although each inspector’s time is chargeable and project scientists need to beware that APHA staff cannot spend extra time at nurseries without explaining to owners why. APHA can provide information on how to package and send project nursery samples up. We also need to make sure that project involvement does not affect APHA’s ISO9000 accreditation.
John Speirs Senior Policy Advisor, Scottish Government;
The Scottish Government has roles from policy to inspectorate and scientific support (through SASA). Though Plant Health is devolved, in general, Plant Health links are strong across the UK. John Speirs is Chair of the Scottish Phytophthora steering group too. A Plant Health Strategy for Scotland was published earlier this year.
John stressed the importance of the project managing its message to industry so that it is viewed positively. John also mentioned the possibility of new EU legislation meaning that nurseries with an ‘action plan’ may not be inspected so frequently.
Richard McIntosh Assistant Chief Plant Health Officer, DEFRA;
Richard is head of the Plant Health Risk and Horizon Scanning team and provides direct support to Nicola Spence, Chief Plant Health Officer as well as scientific and technical training to DEFRA, FERA etc. Richard provided information on the Plant Health Risk Group (PHRG) which meets every 6 months to assess UK wide positions on specific pest issues, including horticultural pest and pathogen problems. The group monitors interception data from the UK and abroad and decides which organisms to place on the UK risk register. Pest Risk Analyses are then commissioned followed by a 12-week consultation period. Recommendations for action are then made to the Chief Plant Health Officer and escalated to ministers where necessary. Currently about 10 species are added to the risk register each month. Richard listed some of the actions required for the 15 Phytophthora species currently on the risk register (11 present in the UK – 4 widespread and 5 more limited in distribution). Richard stated that some challenges to the project include how to prioritise species as threats, dealing with the ‘unknowns’, and the need to offer practical and proportional guidance. In order to secure nursery participation, stakeholders need to know what’s in it for them.
Jon Knight; Head of Research and KT, ADHB
Jon is the Head of Crop Health and Protection at ADHB which is part of DEFRA. It is a levy-raising board and a non-departmental Government Body. Its purpose is to provide independent, evidence-based information and tools for growth and sustainability. 9% of its income (ie about 700k of a total income of £7.2M) comes from the Hardy Nursery Stock sector – 12% of that is from tree production. Phytophthoras cross several sectors of AHDB so is of much interest to them. Jon can help with project in terms of providing stakeholder contacts as he has a list of 600-700 Hardy Nursery Stock sector levy payers. Jon’s presentation summarised the value of the different sectors of the UK horticulture and landscaping industries including the value placed on tourist visits to UK parks and gardens, the amount (£1.4 bn) spent by tourists in gardens, the £2 bn value of UK flower/plant production, and 300k people employed in horticulture and landscaping in the UK. He also mentioned the Ornamental Horticulture Roundtable Action Plan as being of interest to the project which includes Plant Health as a focus.
Phyto-threats workshop 4 October
October 4th 2017, APHA, Sand Hutton, York
Sarah Green, Phyto-threats project co-ordinator, welcomed everyone and introduced the aims of the workshop. These were to:
- share key science findings from the Phyto-threats project which might help underpin accreditation
- understand existing UK assurance schemes and how they might be supported
- generate ideas for how an accreditation scheme should work in order to be effective
The meeting was attended by c50 nursery managers, Plant Health inspectors, foresters, academics, policymakers and others. This report provides an overview of the presentations given on the project team’s research; existing and emerging schemes (UK Sourced and Grown scheme and HTA’s pilot project); and Defra’s position on accreditation. Slides accompanying these talks are available on the Phyto-threats project website (see link above). The report also includes the outcome of discussions when attendees were asked to share thoughts and experiences on ‘how to give accreditation teeth’.
1.1 RESEARCH HIGHLIGHTS
Sarah Green (Forest Research) welcomed the delegates and provided an overview of the project, reiterating the aim to address global threats from Phytophthora species, and to mitigate disease through nursery best practice. The progress of the project to date was summarised, and it was stressed that a number of lessons had been learned surrounding appetite for accreditation, and drivers and challenges facing nurseries (both from partner nurseries and from the overseas perspectives of Susan Frankel (USA) and Giles Hardy (Australia) who featured in last year’s workshop). Sarah concluded by outlining the aim of this workshop: to explore how to make accreditation work and how it should be supported. In addition, the workshop served as an opportunity for partners to hear about the specifics of the research implemented over the past year.
Dave Cooke (James Hutton Institute) spoke on the sampling procedures used during his team’s visits to nursery sites, and the subsequent findings the ongoing analyses are yielding. Samples from fifteen partner nurseries have been collected, including 8 nurseries in Scotland, 6 in England and 1 in Wales. In total over 1700 samples have been collected to date. Analysis of these samples is ongoing. Over 400 samples have been PCR tested for Phytophthora (93 from plant roots of 35 different hosts; 132 water filters; and 170 buffer solutions associated with the filters). The analysis is key to understanding which Phytophthora species are found in nurseries and which management practices contribute to spread or mitigation. This information is expected to help inform nurseries which Phytophthora species represent an emerging threat, allowing proactive action to be taken. Early analysis appears to be demonstrating that mud and puddles, unmanaged shelter belts and ‘hospital areas’ for sickly plants all increase the risk of Phytophthora being harboured.
Mike Dunn (Forest Research) summarised the findings of a consumer survey from 1500 UK plant buyers. The results showed that the public have little awareness about the threats from newly introduced pests and diseases, or the specific pathogens already present. Moreover, when choosing where to buy plants, quality, cost and range of plants are the most important drivers. Presence of biosecurity practices and plant provenance are unimportant in comparison. In terms of acquisition, the most relied upon sources are garden centres (used by 80% of the sample), DIY stores (56%), supermarkets (48%), self-grown from seed (47%) and nurseries (36%) – highlighting the value of an accreditation scheme that could encompass more than just nurseries. Data on purchasing behaviour demonstrated that many of the public buy other accredited/certified products (e.g. Fairtrade products) on the grounds that they agree with the ideals of the scheme, but also because the status implies a high quality product. Forty-five percent stated they would be likely to travel further to buy accredited plants (mean distance of 26.2 miles each way), and 39% reported they would be likely to pay an additional premium (mean premium of 18%). It was acknowledged that the public represent one of several different types of consumer for nurseries. Further research into other customers (e.g. landscapers) is ongoing.
Dan Chapman (Centre for Ecology and Hydrology) moved away from thinking about nurseries to look at the national and global scales of plant movement and colonisation. Particular attention was paid to variations in environmental conditions, and the associated implications for the level of risk posed by different Phytophthoras. This process has involved tapping into global databases on trade and looking at reports of Phytophthora introductions, understanding links between traits and impact, and working towards predicting ‘invasiveness’ in the UK (on the basis of traits and the country’s climatic conditions). Pursuing this research may aid in assessing whether a newly discovered or introduced Phytophthora will be problematic. As such, it is relevant for national scale biosecurity planning, and collaborators connected to the UK plant health risk register.
1.2 EXISTING AND EMERGING SCHEMES
Lee Dudley (Woodland trust) described the Woodland Trust’s UK Sourced and Grown (UKSG) assurance scheme relevant to forest nurseries. Prior to the scheme the Trust were ‘spot buying’ plants from mainland Europe, though this was considered a problem in light of the ash dieback outbreaks. After research and face-to-face discussion with nurseries it was decided that contract growing and seed collection would represent a more secure means of attaining healthy trees from within the UK. Nurseries and seed collectors are now encouraged to seek accreditation, and produce assured products for the Trust. Staff training, seed handling, traceability, stock control, biosecurity, plant quality and quantity, care of soil and water, and seed receipts are all assessed as part of the accreditation process. A total of 30 nurseries were approached for inclusion, leading to 19 agreeing to be audited and subsequently passing, and an additional one requiring corrective action. As a result, an estimated 56.8 million plants have been assured. While the contracts are significant to the forest nursery sector (£2-3 million per year), a huge gap is predicted between what is being supplied by forest nurseries and the number of trees that are expected to be needed in the future. It is hoped that more nurseries and seed collectors will be encouraged to join the scheme, which offers a guaranteed market for assured produce and is a good advert for the nursery. Growth of the scheme will continue to reduce the reliance on spot buying.
Tim Edwards (Boningale Nurseries) provided an overview of the HTA Plant Health Assurance Scheme which is now in its pilot stage. The scheme includes 10 nurseries of varying size and type that are currently being audited as a means of testing the proposed standard for the scheme. It was noted that the nurseries participating are likely to be better than average at managing for biosecurity risks having volunteered to be part of the pilot. However, auditors are noting a number of shortcomings such as a lack of risk assessment, the absence of records of disposals, and a need for further staff training. Those involved are said to be pleased with the process which is helping them to become exemplars within the sector – something which customers such as retailers and amenity planters look favourably upon. A project meeting in November 2017 will explore the governance of the scheme and establish an independent entity that will own and oversee a refined standard for future use. All nurseries will be offered the chance to get up to speed for when the scheme rolls out to avoid giving undue advantage to those participating in the pilot scheme.
1.3 DEFRA’s POSITION ON ACCREDITATION
Nicola Spence (Defra’s Chief Plant Health Officer) acknowledged Defra’s interest in accreditation as well as preventative measures, for example through more thorough host inspections (particularly for highly susceptible species). To illustrate the scale of the challenge faced, Nicola referred to the growing risk register, which typically receives 5-10 new additions at each monthly meeting, and emphasised the need for 5 P’s within the trade: predicting, preventing, protecting, preparing and partnering. Defra’s stance is that nursery accreditation could be an important element in shaping the UK as a trusted provider of quality plants (with reduced pest and disease risk) and thus a bigger exporter. Reducing the number of imports and employing measures on those that do still need to be imported (e.g. quarantining) could not only lower biosecurity risk, but also foster financial resilience within the industry. This resilience could be furthered by restructuring of a grant scheme currently operating for Phytophthora and a limited number of other pathogens. Defra and APHA should remain a source of information on plant pests and diseases via the plant information portal. In addition, Nicola agreed with a suggestion that Defra should influence planning and development policy to insist on the use of accredited nurseries or products during developments, once a standard had been agreed upon as to what is a bio-secure plant.
2. How to Give Accreditation Teeth: Uptake, Compliance and Impact
2.1 Accreditation coverage
Need for extensive coverage : In order for an accreditation scheme to have impact it was agreed that it would need to be; endorsed by the government, critical to the business, and to encompass the products/practices of as many actors within the supply chain as possible. This coverage is deemed necessary given that retail outlets received criticism for current practices, such as offering ‘bargain priced plants’ which had been retained so long as to increase the probability that they harboured pathogens – something which would not be tolerated with the sale of food or animals.
Challenges for widespread inclusion: Developing and implementing a scheme capable of maintaining relevance and appeal to all of the actors within the supply chain is considered a key challenge, undermined by a number of factors. UK nurseries were said to be too small to satisfy the enormous customer base resulting in a sizeable market share for non-specialist traders including supermarket and DIY chains. While the establishment of UK cooperatives emerged as a potential means of rolling out accreditation to a larger number of growers, it was noted that at present different actors appear to be pursuing their own approaches rather than uniting towards a single cross-cutting, standardised approach. For example, one DIY store are said to have a new ‘unification scheme’ which may involve selling only their own branded products, similar to the Ikea model. Other groups have sought to improve the quality of plants by using suppliers accredited under the BOPP scheme. While these developments serve to demonstrate that many have come to recognise the value of some form of assurance/accreditation approach, it is possible that the diversity of what is being proposed will lead to confusion among customers.
Building on existing frameworks : Examples given of successfully established, widespread schemes included sustainable timber schemes and pesticide schemes, raising whether lessons could be learned from these, or if there was an opportunity to ‘piggyback’ – adding biosecurity best practice to an existing scheme. It was agreed that ideally the UK should have a single, recognisable assurance scheme and that existing schemes (i.e., UKSG, HTA, BOPP) should be amalgamated, yet how this could be achieved was not clear.
International coverage: While discussions focussed on encouraging accreditation coverage among the different stakeholders in the UK, others felt strongly that a UK wide accreditation scheme would in fact need to be mandatory for all growers if it were ever to be effective. Certain nursery managers and DIY chain representatives went further still, expressing that accreditation should extend not only to countries on the continent – from which the UK receives the majority of its plants – but also to countries outside Europe. This, they believed, was necessary to ensure that the plants arriving to and leaving from European nurseries could be considered “safe”.
2.2 Support needed within the trade
Ensuring demand for accredited plants : Growers investing in enhanced biosecurity practices seek assurance that their products will be in demand. One way to help ensure this is for the government to insist that contracts must specify the need for accredited trees/plants. However, at present Local Authorities are routinely awarding contracts to the lowest bidder, with little to no attention being paid to plant type, quality or health. More generally, contract growing has not been successful because of a tendency for schemes to be put on hold or not to materialise at all. As a result, it can be extremely difficult to match supply with demand. Ideally procurement/contract growing should nominate an accredited supplier to ensure that plants and trees purchased arrive from a site with the necessary biosecurity practices in place. In addition, there should be a guarantee that the plants/trees will be needed and therefore purchased. This would ensure that growers can manage their supply without fear of the demand disappearing at short notice, and losses being incurred.
Benefits for accredited growers : In the event that a contract is terminated or amended (thus reducing the number of plants/trees required) growers would require an insurance policy or compensation to offset any costs invested in producing the order. By making access to this type of insurance scheme available only to those growers that had been accredited, it would be possible to discourage acquisition of plants from riskier pathways typified by the ‘white van man’ – since traders ineligible for the insurance would be subject to wasted resources when a contract is cancelled, and therefore be at a disadvantage in the marketplace. Other suggested benefits to holding accreditation included eligibility to grant funding and access to different markets (i.e. allowing for the purchase of plants from certain places). Again, these measures would potentially put those without accreditation at a disadvantage and reduce their market share.
Insurance providers : While it was suggested that the government may oversee the administration of financial incentives/reimbursement for those with accreditation, there was a degree of cynicism about how likely this would be. Nevertheless, some did view revisions to the government’s post-Brexit budget as an opportunity to introduce such measures. Other insurance providers were also suggested though it was acknowledged that more discussion would be needed to outline the specific circumstances required for reimbursement, and what level of reimbursement would occur. At present insurance policies which reflect the complexities of the trade were said not to exist, prompting some to highlight a need for the insurance sector to adapt.
Consistent support : In addition to a proposed role in reimbursement, there was also consensus that the Government would need to offer wider support for an accreditation scheme, and be consistent in its efforts to encourage its uptake and effectiveness, for example, by outlining a joined up vision between departments (less silo-ing). Consistency should be visible from the national level (e.g. increasing border controls and inspections), down to local governments, who are considered important due to their role in dealing with outbreaks on the ground. Many also noted a desire for the government to act in an educational role through the provision of more information on the cost of outbreaks, to highlight the extent of financial impacts which result.
2.3 Awareness and Education
Importance of awareness and education : Discussions about education and raising awareness were deemed relevant to plant buyers (including the plant buying public, landscapers, and middle-men in the supply chain) and wider society. Without the knowledge of what accreditation is trying to achieve and what it represents, it cannot be expected to garner support or influence spending behaviour.
Government signs : Achieving improved awareness of Plant Health and its importance was seen by many to be the responsibility of the government and its relevant departments (e.g. DEFRA, APHA). To achieve this aim it was agreed that the problems accreditation would attempt to solve must be made visible to the point where they simply cannot be missed or ignored. Suggested strategies for public spaces included signage throughout transport networks, typified elsewhere by the large, unmissable signs (e.g. bordering Canadian highways in and out of at-risk areas). Bus shelters and airport signage were similarly suggested. The imagery used in these instances could include altered landscapes, featuring ‘before and after’ photographs in outbreak areas, or artificial images to demonstrate how a currently valued landscape would likely appear following a loss of trees.
Media campaigns : TV campaigns, comparable to anti-smoking and green cross code advertisements were thought to be the most effective way to bring the message around the need for accreditation into the publics’ homes. This was thought to give the public little choice, but to see and hear the required information. In contrast, online resources (such as videos or factsheets) are thought to have less impact – since people would actively have to seek out this information themselves it is unlikely that a sizeable proportion of the population would benefit from this form of exposure. For both TV and online content there was some concern that cartoons have a lifespan, and that beyond this the content may become ignored or tiresome if not periodically reinvented. Even with an ongoing campaign, there remained some scepticism that awareness would be long-lasting, and that many people would pay little attention to the issue until they themselves were directly impacted – i.e. through receiving diseased plants or recognising degradation or loss in a forest they used (e.g. discolouration or removal of numerous trees).
The role of growers and sellers : In addition to a government led blanket approach to raising awareness, it was agreed that growers and sellers have an important role to play owing to their first-hand interaction with consumers. As well as having signage and staff on-hand to communicate the problems associated with poor biosecurity and the goals of the scheme, it was felt that compliance with accreditation would need to be visible. This task would be best achieved with the presence of a recognisable accreditation logo that featured on accredited premises and products, alongside an explanation of the scheme’s aims and requirements.
Pressure groups : Pressure groups were also suggested as a means of raising the public’s awareness of risks within the trade. Greenpeace’s campaign to raise awareness about the use of neonicotinoids in pest control put pressure on growers to stop using these products, leading some to suggest that similar action could be employed to address the sale of high risk plants.
Message tone and content : In terms of the tone of the information being disseminated, it was suggested that finger pointing be avoided, and instead the focus be on informing about the problem and its solutions. Others felt that there was a need to go beyond informing on the threat to the landscape, and to educate customers on what goes into growing a plant – something which may chime with the large proportion of those whose buying choices are influenced by plant quality. Some went as far as to say that accreditation should include a warranty/guarantee to customers to demonstrate that the product is high-end and worth paying for. Whatever the avenue for engaging with the public, it was deemed important that the message be succinct and trustworthy. Indeed, trust in the accreditation scheme and the growers are considered vital to the public’s receptiveness to supporting the scheme’s goals.
2.4 Training for the trade
Integration of training into accreditation : Early discussions about the posited and emerging accreditation schemes (including the HTA pilot project) have included proposals for ongoing improvement of a nursery’s practices in combination with staff training. Although the HTA pilot nurseries were described as having, “a lot to do, with few provisions” there is some optimism that inroads are being made, with DEFRA being particularly helpful in thinking about how training might be delivered.
Training options: Although the likes of SRUC’s new MSc on forensic plant health were highlighted as potentially valuable courses, they tend to be in short supply. In addition, the workforce of some nurseries are without the qualifications – and perhaps desire – to attain the level of expertise suggested to be necessary, meaning this formal education approach would prove inaccessible or incongruous to many workers. Instead, the idea of having an assigned officer within the organisation (equivalent to a health and safety officer) was proposed. This could be encouraged by making the presence of such a position a compulsory requirement for accreditation.
Knowledge and skills throughout the sector : Some felt that plant health professionals should be embedded in other professions including landscape design and retail. This measure was seen as a means of reducing the promotion and demand for high-risk trees and plants (such as olive trees and Himalayan balsam). It would also presumably help move towards a sector-wide commitment to selling only those plants which are in season, and more generally, facilitating sound biosecurity becoming revered throughout the supply chain. The upcoming introduction of a training module by the Landscape Institute is one example where increasing knowledge of plant health and best practice will soon be encouraged. Meanwhile in Scotland, the Confor nursery group is looking to see how to offer and deliver training on plant health to growers.
The rewards of training : Finally, it was suggested that the best practice and training required for accreditation should be something which is promoted to the growers on the grounds that a better quality product would result. On this basis, accreditation may become an asset (a badge of honour for growers and sellers), rather than representing red tape or a tick box exercise.
2.5 Challenges
Added costs : One nursery manager queried whether accreditation would necessitate more costs for the consumer – something which may undermine a scheme and accredited businesses should plant buyers opt to go elsewhere in search of cheaper goods. They reasoned that if implemented efficiently within nurseries, biosecurity should not be excessively costly and thus the costs passed on to consumers would be negligible. However, others with knowledge of schemes elsewhere insisted that it would be impossible for many nurseries to enact management changes and improved biosecurity measures without increasing the price of its products. In addition, it was noted that any input from plant health inspections would be expensive to growers, since inspectors charge even for 15 minutes of their time. Therefore, resources need to be put in place so that a scheme can be policed without excessively burdening growers. This policing may involve inspectors and auditors being granted the power to force changes on nurseries. The idea of a yellow/red card system for those who have failed to comply was also suggested, yet the specifics about whether non-compliance would result in any loss of privileges or the incurrence of fines – and over what timeframe – require further discussion.
Limitations to accreditation : Finally, it was acknowledged that an accreditation scheme could only be expected to reduce the risk of pathogen spread as opposed to eliminate it. For this reason, some raised concerns about the robustness of a scheme’s credibility in the event of an outbreak, particularly if it were to occur in the early days of a scheme being rolled out. As it is impossible to discount this possibility, the participant cautioned against overselling what the accreditation scheme could achieve.
2.6 KEY OUTCOMES
- A single, all-encompassing UK accreditation scheme is preferred
- Accreditation needs to cover the entire supply chain if it is to have impact
- Demand for accredited products should be ensured to incentivise uptake in the scheme
- Accreditation could also be incentivised by allowing access to grant funding, and through financial reimbursement when contracts to supply accredited plants are cancelled/altered
- It is necessary to raise public awareness about the need and benefits of accreditation through highly visible campaigns and signage, including a recognisable logo
- Training for growers and others in the sector should be integral to accreditation so that practices continue to improve
- For a scheme to be respected and to generate improvement it must be effectively policed
Research Forum on Invasive Species
Phyto-threats attendance at the 28th USDA Interagency Research Forum on Invasive Species held in Annapolis, Maryland, USA, January 10-13, 2017
Sarah Green (Forest Research) attended the 28th USDA Interagency Research Forum on Invasive Species held in Annapolis, Maryland, January 10-13, 2017. She presented a paper entitled ‘Tackling emerging forest Phytophthoras in the UK: Mitigating risk of new introductions and managing diseased landscapes for the future’ during the session ‘Forest Phytophthora: They get around’ organized by IUFRO Working Party 7.02.09, Phytophthoras in Forests and Natural Ecosystems. Sarah’s presentation included an overview of the LWEC3 Phyto-threats project; rationale and objectives. The talk generated discussion on the overall willingness of nurseries to participate in the project, and the conundrum posed by ever increasing trade flows whilst trying to reduce risks of global spread of pests and diseases. Sarah also took the opportunity to ask for collaborators to assist the Phyto-threats workpackage 3 team, led by Beth Purse, in compiling data on global Phytophthora occurrence. She learned of an ongoing project by Yilmaz Balci from USDA-APHIS who has about 21 new Phytophthora species (as yet undescribed) which he will publish on this year. Twelve of the species were collected during surveys in South and Central America and the rest were collected from eastern USA. These will be added to the Phyto-threats traits’ database when the data become available. In general there was much enthusiasm for having one central Phytophthora database (listing biological traits and distribution) available globally. This is a big project however and one which would require additional resources to manage beyond the lifetime of Phyto-threats.
There were a number of other speakers who gave presentations of particular interest to the Phyto-threats project. Everett Hansen of Oregon State University reported on the high diversity of Phytophthoras found during surveys of Oregon wildlands. Laura Sims, University of California, discussed how plant disease predictions for invasive soilborne Phytophthora species are consistent with host ecology and with genus-level co-evolutionary history. Rebecca Epanchin-Niell of ‘Resources for the Future’ based in Washington DC described a cost-benefit analysis of the live plant trade during her talk ‘Informing efficient strategies to reduce pest risk from live plant imports’. The analysis was done in relation to risks to US forests from introductions of insect pests, but it involved economic estimates of the welfare benefits of the live plant trade, expected damages per forest pest establishment (over time as invasion spreads) and included an assessment of the relatedness of imported plant species to important forest species in the US. This work can be viewed in more detail in; Epanchin-Niell and Liebhold, 2015. Benefits of invasion prevention: Effect of time lags, spread rates, and damage persistence. Ecological Economics 116; 146-153
Also of interest was a talk by Rebecca Ganley of Scion, Rotorua, New Zealand, who described her colleague Peter Scott’s work in compiling a list of all Phytophthora species reported in every country globally. They are using modelling approaches to predict the number of Phytophthora species likely to be present in each country in the world. The Phyto-threats workpackage 3 team are now in touch with the NZ group to scope the potential for collaboration/sharing of resources.
Research partners
 |
 |
|
 |
||
 |
Forest Research (FR)
Beatrice Henricot
James Hutton Institute (JHI)
Centre for Ecology and Hydrology (CEH)
Bethan Purse
University of Edinburgh
Eden Project (previously University of Worcester)
Animal and Plant Health Agency (APHA)
Jane Barbrook
Science and Advice for Scottish Agriculture (SASA)
Alexandra Schlenzig
